1RW1
 
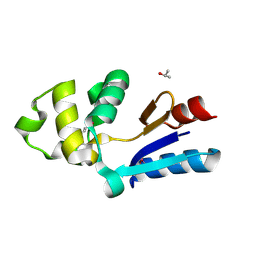 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1YI7
 
 | | Beta-d-xylosidase (selenomethionine) XYND from Clostridium Acetobutylicum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-xylosidase from clostridium acetobutylicum
To be Published
|
|
1Y7B
 
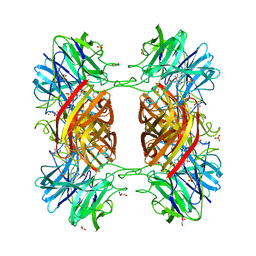 | | BETA-D-XYLOSIDASE, A FAMILY 43 GLYCOSIDE HYDROLASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-xylosidase, family 43 glycosyl hydrolase, ... | | Authors: | Teplyakov, A, Fedorov, E, Gilliland, G.L, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-08 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of beta-xylosidase from Clostridium acetobutylicum
To be Published
|
|
3L7E
 
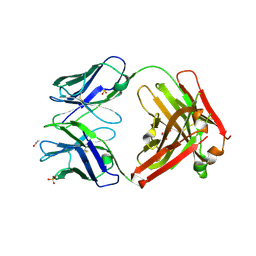 | | Crystal structure of ANTI-IL-13 antibody C836 | | Descriptor: | ACETATE ION, C836 HEAVY CHAIN, C836 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen recognition by antibody C836 through adjustment of VL/VH packing
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5KNG
 
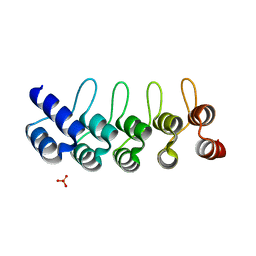 | | CRYSTAL STRUCTURE OF ANTI-IL-13 DARPIN 6G9 | | Descriptor: | DARPIN 6G9, GLYCEROL, PHOSPHATE ION | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
5KNH
 
 | | CRYSTAL STRUCTURE OF DARPIN 6G9 IN COMPLEX WITH CYNO IL-13 | | Descriptor: | ACETATE ION, DARPIN 6G9, IL13 | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Gilliland, G. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational flexibility of an anti-IL-13 DARPin.
Protein Eng. Des. Sel., 30, 2017
|
|
1NOA
 
 | |
1R4B
 
 | |
1R45
 
 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
1JXA
 
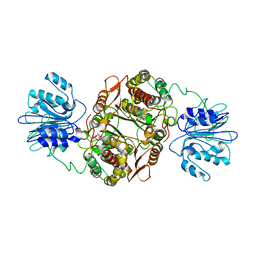 | | GLUCOSAMINE 6-PHOSPHATE SYNTHASE WITH GLUCOSE 6-PHOSPHATE | | Descriptor: | GLUCOSE-6-PHOSPHATE, glucosamine 6-phosphate synthase | | Authors: | Teplyakov, A, Obmolova, G, Badet, B, Badet-Denisot, M.A. | | Deposit date: | 2001-09-06 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Channeling of ammonia in glucosamine-6-phosphate synthase.
J.Mol.Biol., 313, 2001
|
|
4LPV
 
 | | Crystal structure of TENCON variant P41BR3-42 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TENCON variant P41BR3-42 | | Authors: | Teplyakov, A, Obmolova, G, Luo, J, Gilliland, G.L. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal beta-strand swapping in a consensus-derived fibronectin Type III scaffold.
Proteins, 82, 2014
|
|
4LPT
 
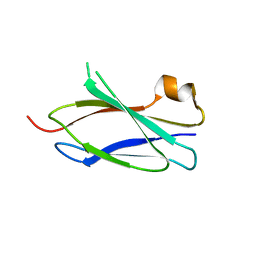 | |
4LPW
 
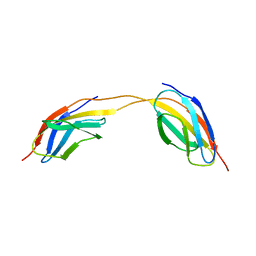 | |
4LPY
 
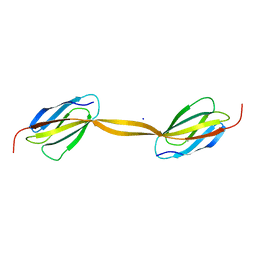 | |
4LPU
 
 | |
4LPX
 
 | |
1JOP
 
 | |
1M65
 
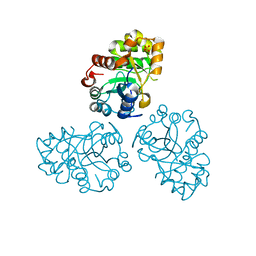 | | YCDX PROTEIN | | Descriptor: | Hypothetical protein ycdX, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-07-12 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1M68
 
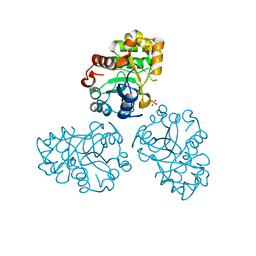 | | YCDX PROTEIN, TRINUCLEAR ZINC SITE | | Descriptor: | Hypothetical protein ycdX, SULFATE ION, ZINC ION | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L. | | Deposit date: | 2002-07-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a
trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1JAL
 
 | |
1HTW
 
 | |
1FL9
 
 | | THE YJEE PROTEIN | | Descriptor: | HYPOTHETICAL PROTEIN HI0065, MERCURY (II) ION | | Authors: | Teplyakov, A, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2000-08-13 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the YjeE protein from Haemophilus influenzae: a putative Atpase involved in cell wall synthesis
Proteins, 48, 2002
|
|
1IN0
 
 | |
4M6O
 
 | | Crystal structure of anti-NGF antibody CNTO7309 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO7309 heavy chain, CNTO7309 light chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M7K
 
 | | Crystal structure of anti-tissue factor antibody 10H10 | | Descriptor: | 10H10 heavy chain, 10H10 light chain, ACETATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
