1F75
 
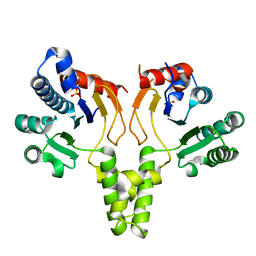 | | CRYSTAL STRUCTURE OF UNDECAPRENYL DIPHOSPHATE SYNTHASE FROM MICROCOCCUS LUTEUS B-P 26 | | Descriptor: | SULFATE ION, UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | Authors: | Fujihashi, M, Zhang, Y.-W, Higuchi, Y, Li, X.-Y, Koyama, T, Miki, K. | | Deposit date: | 2000-06-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5AVD
 
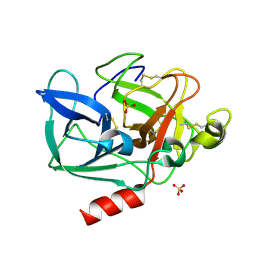 | | The 0.86 angstrom structure of elastase crystallized in high-strength agarose hydrogel | | Descriptor: | Chymotrypsin-like elastase family member 1, SULFATE ION | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVG
 
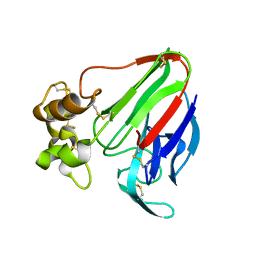 | | The 0.95 angstrom structure of thaumatin crystallized in high-strength agarose hydrogel | | Descriptor: | Thaumatin-1 | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Hirose, M, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5XJA
 
 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF2C, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2017-04-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
5XJB
 
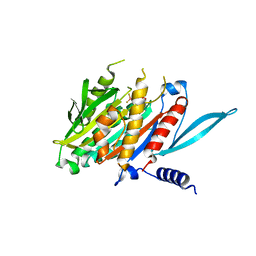 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Kinesin-like protein KIF2C, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2017-04-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
6A97
 
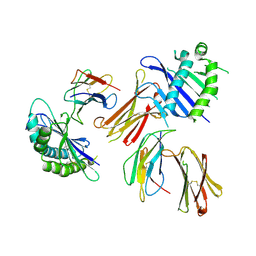 | | Crystal structure of MHC-like MILL2 | | Descriptor: | Beta-2-microglobulin, MHC I-like leukocyte 2 long form, SULFATE ION | | Authors: | Kajikawa, M, Ose, T, Maenaka, K. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-05 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Structure of MHC class I-like MILL2 reveals heparan-sulfate binding and interdomain flexibility.
Nat Commun, 9, 2018
|
|
2ZOI
 
 | |
2ZOH
 
 | |
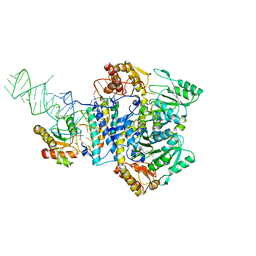
5X6B
 
 | |
5XW7
 
 | |
5X6C
 
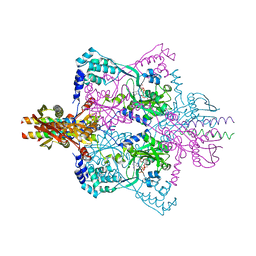 | | Crystal structure of SepRS-SepCysE from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, O-phosphoserine--tRNA(Cys) ligase, SULFATE ION, ... | | Authors: | Chen, M, Kato, K, Yao, M. | | Deposit date: | 2017-02-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural basis for tRNA-dependent cysteine biosynthesis
Nat Commun, 8, 2017
|
|
2Z6D
 
 | |
2Z6C
 
 | |
