6GHL
 
 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, MALONATE ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GFP
 
 | | cyanobacterial GAPDH with NADP bound | | Descriptor: | FORMIC ACID, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Briggs, L, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GFR
 
 | | cyanobacterial GAPDH with NAD | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GG7
 
 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GFO
 
 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GHR
 
 | | cyanobacterial GAPDH with full-length CP12 | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GFQ
 
 | | cyanobacterial GAPDH with NAD and CP12 bound | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GVE
 
 | | GAPDH-CP12-PRK complex | | Descriptor: | CP12 polypeptide, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | McFarlane, C.R, Shah, N, Bubeck, D, Murray, J.W. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HCX
 
 | | Influenza Virus N9 Neuraminidase A complex with Zanamivir molecule (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
4ZGS
 
 | | Identification of the pyruvate reductase of Chlamydomonas reinhardtii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative D-lactate dehydrogenase | | Authors: | Burgess, S.J, Hussein, T, Yeoman, J.A, Iamshanova, O, Boehm, M, Bundy, J, Bialek, W, Murray, J.W, Nixon, P.J. | | Deposit date: | 2015-04-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Identification of the Elusive Pyruvate Reductase of Chlamydomonas reinhardtii Chloroplasts.
Plant Cell.Physiol., 57, 2016
|
|
5A5L
 
 | | Structure of dual function FBPase SBPase from Thermosynechococcus elongatus | | Descriptor: | 7-O-phosphono-alpha-L-galacto-hept-2-ulopyranose, D-FRUCTOSE 1,6-BISPHOSPHATASE CLASS 2/SEDOHEPTULOSE 1,7-BISPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Cotton, C.A.R, Kabasakal, B, Miah, N, Murray, J.W. | | Deposit date: | 2015-06-19 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the Dual-Function Fructose-1,6/Sedoheptulose-1, 7-Bisphosphatase from Thermosynechococcus Elongatus Bound with Sedoheptulose-7-Phosphate.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5ABR
 
 | |
6SDM
 
 | | NADH-dependent variant of TBADH | | Descriptor: | NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
6SCH
 
 | | NADH-dependent variant of CBADH | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
6TVI
 
 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
6TRG
 
 | | Salmonella typhimurium neuraminidase mutant (D100S) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D100S)
To Be Published
|
|
2XBG
 
 | |
2Y6X
 
 | | Structure of Psb27 from Thermosynechococcus elongatus | | Descriptor: | CHLORIDE ION, PHOTOSYSTEM II 11 KD PROTEIN | | Authors: | Michoux, F, Takasaka, K, Boehm, M, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Psb27 Assembly Factor at 1.6A: Implications for Binding to Photosystem II.
Photosynth.Res., 110, 2012
|
|
5LYP
 
 | | Crystal structure of the Tpr Domain of Sgt2. | | Descriptor: | BORATE ION, Small glutamine-rich tetratricopeptide repeat-containing protein 2 | | Authors: | Krysztofinska, E.M, Morgan, R.M.L, Thapaliya, A, Evans, N.J, Murray, J.W, Martinez-Lumbreras, S, Isaacson, R.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Front Mol Biosci, 4, 2017
|
|
5LYN
 
 | | Structure of the Tpr Domain of Sgt2 in complex with yeast Ssa1 peptide fragment | | Descriptor: | PRO-THR-VAL-GLU-GLU-VAL-ASP, Small glutamine-rich tetratricopeptide repeat-containing protein 2, ZINC ION | | Authors: | Krysztofinska, E.M, Evans, N.J, Thapaliya, A, Murray, J.W, Morgan, R.M.L, Martinez-Lumbreras, S, Isaacson, R.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Front Mol Biosci, 4, 2017
|
|
3ZSU
 
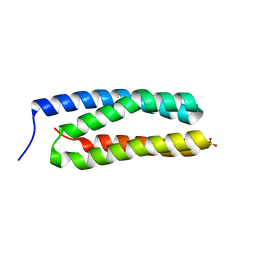 | | Structure of the CyanoQ protein from Thermosynechococcus elongatus | | Descriptor: | SULFATE ION, TLL2057 PROTEIN | | Authors: | Michoux, F, Takasaka, K, Nixon, P.J, Murray, J.W. | | Deposit date: | 2011-06-30 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Cyanoq from the Thermophilic Cyanobacterium Thermosynechococcus Elongatus and Detection in Isolated Photosystem II Complexes.
Photosynth.Res., 122, 2014
|
|
3ZPN
 
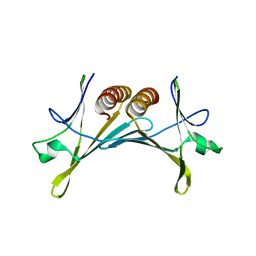 | | Structure of Psb28 | | Descriptor: | PHOTOSYSTEM II REACTION CENTER PSB28 PROTEIN | | Authors: | Bialek, W.J, Michoux, F, Nixon, P.J, Murray, J.W. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Crystal Structure of the Psb28 Accessory Factor of Thermosynechococcus Elongatus Photosystem II at 2.3 A
Photosynth.Res., 117, 2013
|
|
3ZCX
 
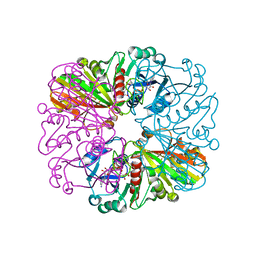 | |
3ZDF
 
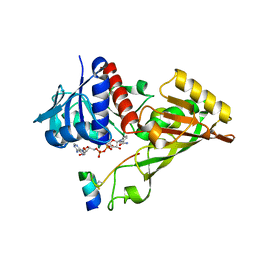 | |
5FRT
 
 | |
