6SNH
 
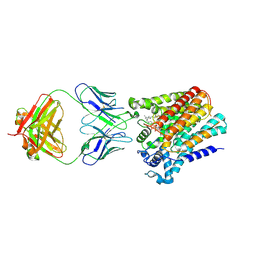 | | Cryo-EM structure of yeast ALG6 in complex with 6AG9 Fab and Dol25-P-Glc | | Descriptor: | 6AG9 Fab heavy chain, 6AG9 Fab light chain, Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase, ... | | Authors: | Bloch, J.S, Pesciullesi, G, Boilevin, J, Nosol, K, Irobalieva, R.N, Darbre, T, Aebi, M, Kossiakoff, A.A, Reymond, J.L, Locher, K.P. | | Deposit date: | 2019-08-24 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and mechanism of the ER-based glucosyltransferase ALG6.
Nature, 579, 2020
|
|
7A65
 
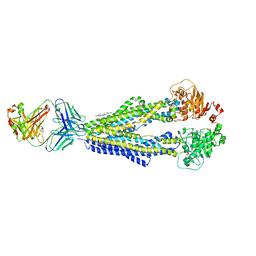 | |
7A6E
 
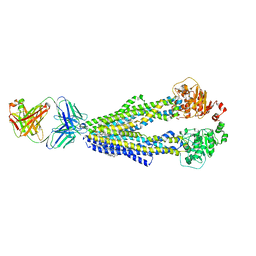 | |
7A69
 
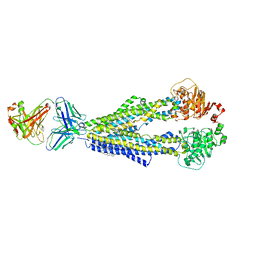 | |
7A6F
 
 | |
7A6C
 
 | |
6S7P
 
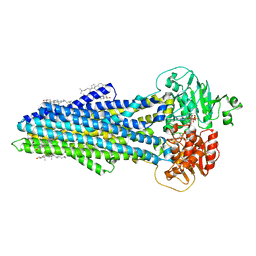 | | Nucleotide bound ABCB4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Olsen, J.A, Alam, A, Kowal, J, Stieger, B, Locher, K.P. | | Deposit date: | 2019-07-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human lipid exporter ABCB4 in a lipid environment.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5OVW
 
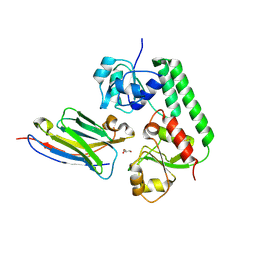 | | Nanobody-bound BtuF, the vitamin B12 binding protein in Escherichia coli | | Descriptor: | GLYCEROL, Nanobody, Vitamin B12-binding protein | | Authors: | Mireku, S.A, Sauer, M.M, Glockshuber, R, Locher, K.P. | | Deposit date: | 2017-08-30 | | Release date: | 2017-11-08 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD.
Sci Rep, 7, 2017
|
|
5NJ3
 
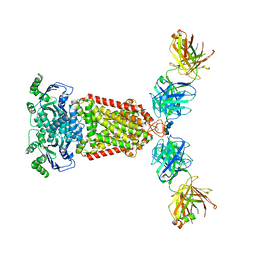 | | Structure of an ABC transporter: complete structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3-Fab heavy chain, 5D3-Fab light chain, ... | | Authors: | Taylor, N.M.I, Manolaridis, I, Jackson, S.M, Kowal, J, Stahlberg, H, Locher, K.P. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
5NO0
 
 | |
5NRP
 
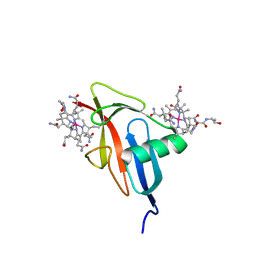 | |
5NP4
 
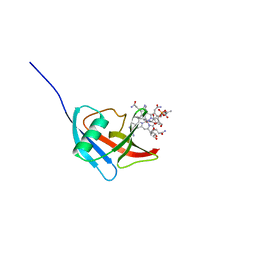 | |
5OGL
 
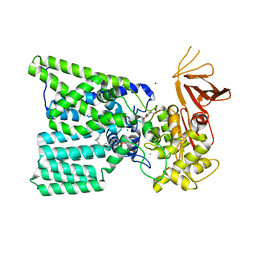 | | Structure of bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Substrate mimicking peptide, ... | | Authors: | Napiorkowska, M, Boilevin, J, Sovdat, T, Darbre, T, Reymond, J.-L, Aebi, M, Locher, K.P. | | Deposit date: | 2017-07-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NSA
 
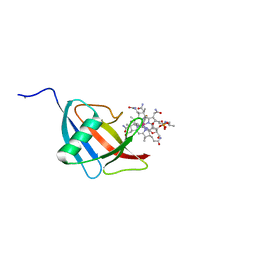 | | Beta domain of human transcobalamin bound to Co-beta-[2-(2,4-difluorophenyl)ethinyl]cobalamin | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, CALCIUM ION, COBALAMIN, ... | | Authors: | Bloch, J.S, Ruetz, M, Krautler, B, Locher, K.P. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.273 Å) | | Cite: | Structure of the human transcobalamin beta domain in four distinct states.
PLoS ONE, 12, 2017
|
|
5NIV
 
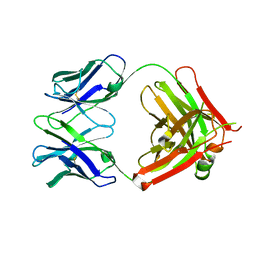 | | Crystal structure of 5D3 Fab | | Descriptor: | Heavy chain of 5D3 Fab, Light chain of 5D3 Fab | | Authors: | Manolaridis, I, Locher, K.P. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of the human multidrug transporter ABCG2.
Nature, 546, 2017
|
|
7OZ1
 
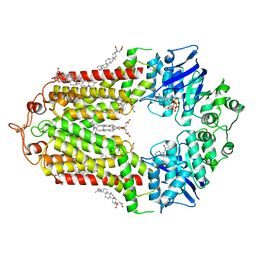 | | Cryo-EM structure of ABCG1 E242Q mutant with ATP and cholesteryl hemisuccinate bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Skarda, L, Kowal, J, Locher, K.P. | | Deposit date: | 2021-06-25 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Human Cholesterol Transporter ABCG1.
J.Mol.Biol., 433, 2021
|
|
4DBL
 
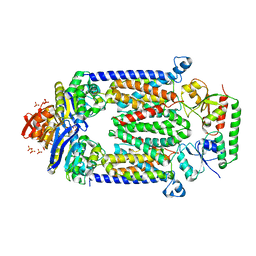 | | Crystal structure of E159Q mutant of BtuCDF | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.M, Hvorup, R.N, Locher, K.P. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-07 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Asymmetric states of vitamin B12 transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF.
Febs Lett., 586, 2012
|
|
5C76
 
 | |
4ZRP
 
 | | TC:CD320 | | Descriptor: | CALCIUM ION, CD320 antigen, CYANOCOBALAMIN, ... | | Authors: | Alam, A, Locher, K.P. | | Deposit date: | 2015-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of transcobalamin recognition by human CD320 receptor.
Nat Commun, 7, 2016
|
|
5C73
 
 | |
5C78
 
 | |
4ZRQ
 
 | |
7NIU
 
 | |
7NIV
 
 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment light chain, CHOLESTEROL, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NIW
 
 | | Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 4B1 Fab-fragment heavy chain, 4B1 Fab-fragment light chain, ... | | Authors: | Nosol, K, Locher, K.P. | | Deposit date: | 2021-02-14 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of ABCB4 provide insight into phosphatidylcholine translocation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
