2EWU
 
 | |
3VXD
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889, SULFATE ION | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
6K31
 
 | |
5B0U
 
 | |
7E9U
 
 | | Trehalase of Arabidopsis thaliana | | Descriptor: | GLYCEROL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7E9X
 
 | | Trehalase of Arabidopsis thaliana acid mutant -D380A | | Descriptor: | GLYCEROL, Trehalase | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
7EAW
 
 | | Trehalase of Arabidopsis thaliana acid mutant -D380A trehalose complex | | Descriptor: | GLYCEROL, Trehalase, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Taguchi, Y, Saburi, W, Yu, J, Imai, R, Yao, M, Mori, H. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH-dependent alteration of substrate specificity of plant trehalase and its molecular mechanism
To Be Published
|
|
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
3WIW
 
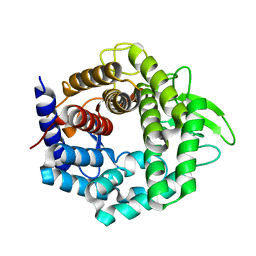 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3WVX
 
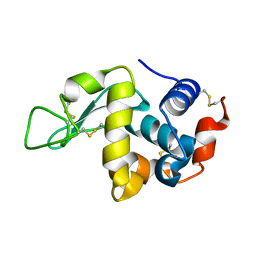 | |
3WVY
 
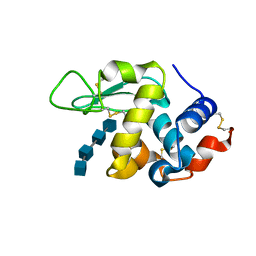 | | Structure of D48A hen egg white lysozyme in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Kawaguchi, Y, Yoneda, K, Araki, T. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The role of Asp48 in the hydrogen bonding network involving Asp52 of hen egg white lysozyme
TO BE PUBLISHED
|
|
6KRN
 
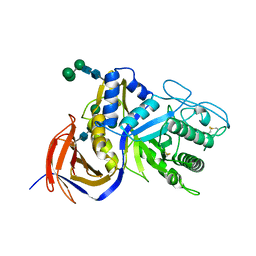 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, Mating factor alpha,GH30 Xylanase B, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6KRL
 
 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6M5Z
 
 | | Catalytic domain of GH30 xylanase C from Talaromyces cellulolyticus | | Descriptor: | ACETATE ION, GH30 Xylanase C, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2020-03-12 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of GH30-7 endoxylanase C from the filamentous fungus Talaromyces cellulolyticus.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5SIC
 
 | |
2TLD
 
 | | CRYSTAL STRUCTURE OF AN ENGINEERED SUBTILISIN INHIBITOR COMPLEXED WITH BOVINE TRYPSIN | | Descriptor: | STREPTOMYCES SUBTILISIN INHIBITOR (SSI), TRYPSIN | | Authors: | Mitsui, Y, Takeuchi, Y, Nonaka, T, Nakamura, K.T. | | Deposit date: | 1991-09-16 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
2RDV
 
 | |
2SIC
 
 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | Authors: | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | Deposit date: | 1991-04-01 | | Release date: | 1993-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
5H09
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-ethyl2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoate | | Descriptor: | Tyrosine-protein kinase HCK, ethyl (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanoate | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0B
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0H
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,N,4-trimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},~{N},4-trimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0E
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0G
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,4-dimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},4-dimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
2YXV
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
2YXW
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
