5LXV
 
 | |
4N2O
 
 | | Structure of a novel autonomous cohesin protein from Ruminococcus flavefaciens | | Descriptor: | Autonomous cohesin, CHLORIDE ION | | Authors: | Frolow, F, Voronov-Goldman, M, Levy-Assaraf, M, Lamed, R, Bayer, E, Shimon, L. | | Deposit date: | 2013-10-05 | | Release date: | 2013-12-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural characterization of a novel autonomous cohesin from Ruminococcus flavefaciens.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4IU2
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cell-wall anchoring protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4IU3
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Cellulose-binding protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4UYP
 
 | | High resolution structure of the third cohesin ScaC in complex with the ScaB dockerin with a mutation in the N-terminal helix (IN to SI) from Acetivibrio cellulolyticus displaying a type I interaction. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Cameron, K, Alves, V.D, Bule, P, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Cell-surface Attachment of Bacterial Multienzyme Complexes Involves Highly Dynamic Protein-Protein Anchors.
J. Biol. Chem., 290, 2015
|
|
4UYQ
 
 | |
6KGC
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 5.4 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGD
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 8.0 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGE
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 5.5 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KG8
 
 | | Solution structure of CaCohA2 from Clostridium acetobutylicum | | Descriptor: | Probably cellulosomal scaffolding protein, secreted cellulose-binding and cohesin domain | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
8X39
 
 | |
8X3A
 
 | |
1NBC
 
 | | BACTERIAL TYPE 3A CELLULOSE-BINDING DOMAIN | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A | | Authors: | Tormo, J, Lamed, R, Steitz, T.A. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose.
EMBO J., 15, 1996
|
|
8HDJ
 
 | |
8HER
 
 | |
8HEP
 
 | |
8HEQ
 
 | |
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I24
 
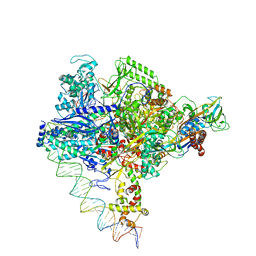 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
6HDS
 
 | | Crystal Structure of apo short afifavidin | | Descriptor: | short afifavidin | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2018-08-19 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of afifavidin reveals common features of molecular assemblage in the bacterial dimeric avidins.
FEBS J., 285, 2018
|
|
6IVU
 
 | |
6IVS
 
 | |
6HDV
 
 | |
6KG9
 
 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
