3WFU
 
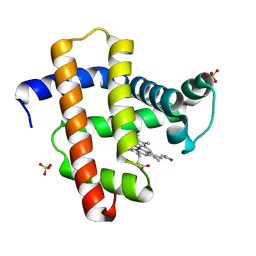 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(I) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, Myoglobin, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
3WJG
 
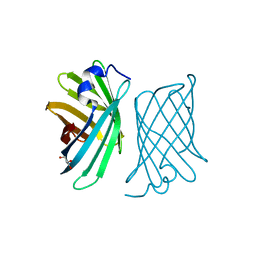 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148W/H158L (NB10) from Arabidopsis thaliana | | Descriptor: | GLYCEROL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WFT
 
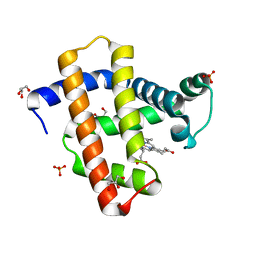 | | Crystal structure of horse heart myoglobin reconstituted with cobalt(II) tetradehydrocorrin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, GLYCEROL, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Hirata, K, Ohbayashi, J, Inoue, T, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2013-07-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Co(II)/Co(I) reduction-induced axial histidine-flipping in myoglobin reconstituted with a cobalt tetradehydrocorrin as a methionine synthase model.
Chem.Commun.(Camb.), 50, 2014
|
|
3WI8
 
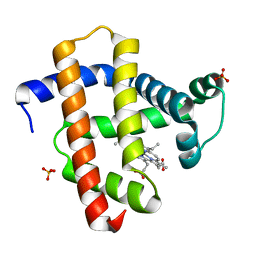 | | Crystal structure of horse heart myoglobin reconstituted with manganese porphycene | | Descriptor: | Myoglobin, PORPHYCENE CONTAINING MN, SULFATE ION | | Authors: | Mizohata, E, Oohora, K, Kihira, Y, Inoue, T, Hayashi, T. | | Deposit date: | 2013-09-09 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | C(sp3)-H bond hydroxylation catalyzed by myoglobin reconstituted with manganese porphycene.
J.Am.Chem.Soc., 135, 2013
|
|
3WJC
 
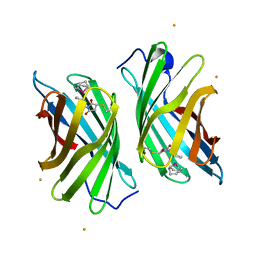 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148L/H158L covalently linked with [Rh(Cp-Mal)(COD)] (NB4-Rh) from Arabidopsis thaliana | | Descriptor: | BARIUM ION, UPF0678 fatty acid-binding protein-like protein At1g79260, [(1,2,5,6-eta)-cyclooctane-1,2,5,6-tetrayl]{(1,2,3,4,5-eta)-1-[2-(2,5-dioxopyrrolidin-1-yl)ethyl]cyclopentadienyl}rhodium | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WJF
 
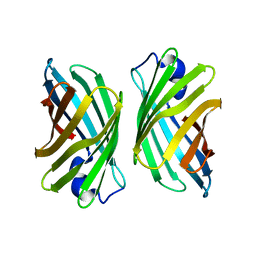 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/V128W/M148L/H158L (NB9) from Arabidopsis thaliana | | Descriptor: | UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
1IWA
 
 | | RUBISCO FROM GALDIERIA PARTITA | | Descriptor: | SULFATE ION, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit | | Authors: | Okano, Y, Mizohata, E, Xie, Y, Matsumura, H, Sugawara, H, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2002-04-30 | | Release date: | 2003-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Galdieria Rubisco Complexed with one sulfate ion per active site
FEBS LETT., 527, 2002
|
|
4TKB
 
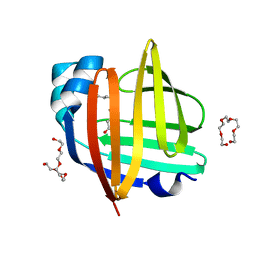 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lauric acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TJZ
 
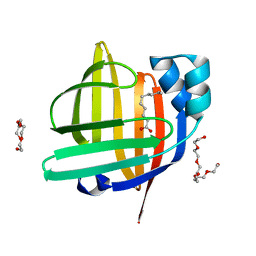 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with capric acid | | Descriptor: | DECANOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKH
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with myristic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKJ
 
 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7XON
 
 | |
7XOM
 
 | |
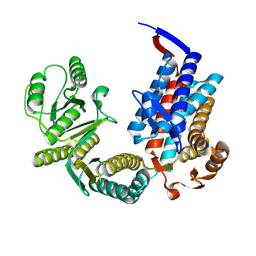
7XOK
 
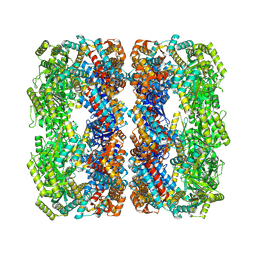 | |
7XOL
 
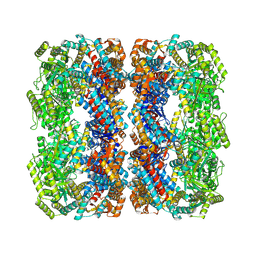 | |
7XOP
 
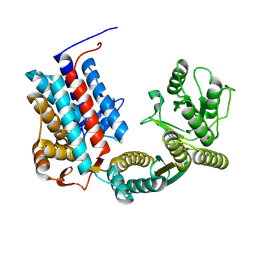 | |
7XOQ
 
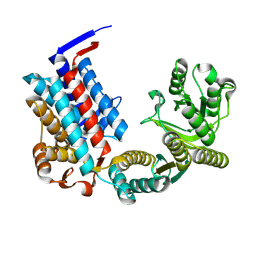 | |
7XOO
 
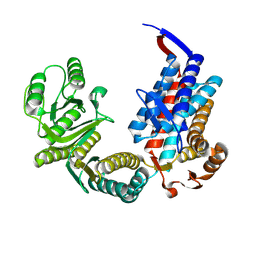 | |
7XOS
 
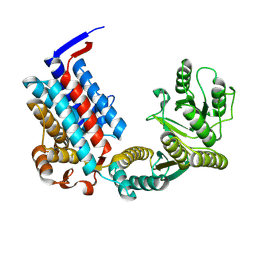 | |
7XOR
 
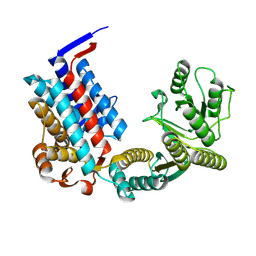 | |
7XOJ
 
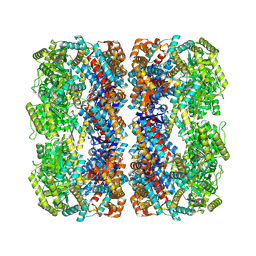 | |
2ZMF
 
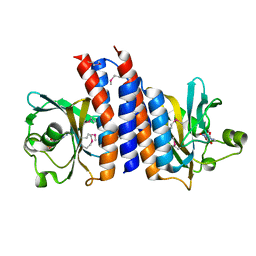 | | Crystal structure of the C-terminal GAF domain of human phosphodiesterase 10A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Handa, N, Kishishita, S, Mizohata, E, Omori, K, Kotera, J, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the GAF-B Domain from Human Phosphodiesterase 10A Complexed with Its Ligand, cAMP
J.Biol.Chem., 283, 2008
|
|
4YST
 
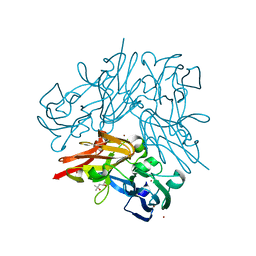 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
3AGT
 
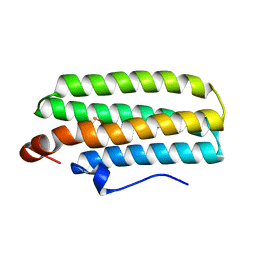 | | Hemerythrin-like domain of DcrH (met) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
3AGU
 
 | | Hemerythrin-like domain of DcrH (semimet-R) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
