3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
1N8N
 
 | | Crystal structure of the Au3+ complex of AphA class B acid phosphatase/phosphotransferase from E. coli at 1.69 A resolution | | Descriptor: | Class B acid phosphatase, GOLD 3+ ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2002-11-21 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
3HQC
 
 | | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Do, J, Dickey, M, Bain, K, Gheyi, T, Klemke, R, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Phosphotyrosine-binding domain from the Human Tensin-like C1 domain-containing phosphatase (TENC1)
To be Published
|
|
7RGF
 
 | | Protocadherin gammaC4 EC1-4 crystal structure disrupted trans interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
3E03
 
 | | Crystal structure of a putative dehydrogenase from Xanthomonas campestris | | Descriptor: | CALCIUM ION, Short chain dehydrogenase | | Authors: | Sampathkumar, P, Wasserman, S, Rutter, M, Hu, S, Bain, K, Rodgers, L, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-09-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of a putative dehydrogenase from Xanthomonas campestris
To be Published
|
|
2KSS
 
 | | NMR structure of Myxococcus xanthus antirepressor CarS1 | | Descriptor: | Carotenogenesis protein carS | | Authors: | Jimenez, M, Gonzalez, C, Padmanabhan, S, Leon, E, Navarro-Aviles, G, Elias-Arnanz, M. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Nucleic Acids Res., 38, 2010
|
|
2GPM
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3') | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1D8H
 
 | | X-RAY CRYSTAL STRUCTURE OF YEAST RNA TRIPHOSPHATASE IN COMPLEX WITH SULFATE AND MANGANESE IONS. | | Descriptor: | MANGANESE (II) ION, SULFATE ION, mRNA TRIPHOSPHATASE CET1 | | Authors: | Lima, C.D, Wang, L.K, Shuman, S. | | Deposit date: | 1999-10-24 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus.
Cell(Cambridge,Mass.), 99, 1999
|
|
2GQ7
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1NBO
 
 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
1N9K
 
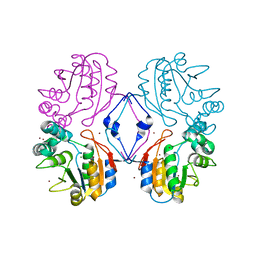 | | Crystal structure of the bromide adduct of AphA class B acid phosphatase/phosphotransferase from E. coli at 2.2 A resolution | | Descriptor: | BROMIDE ION, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2002-11-25 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
6HNC
 
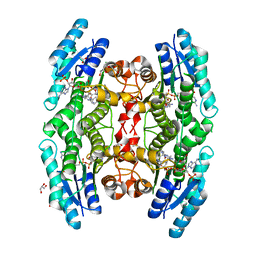 | | Trypanosoma brucei PTR1 in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Development of Cycloguanil Derivatives asTrypanosoma bruceiPteridine-Reductase-1 Inhibitors.
Acs Infect Dis., 5, 2019
|
|
3DOU
 
 | | Crystal structure of methyltransferase involved in cell division from thermoplasma volcanicum gss1 | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYLMETHIONINE | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Hu, S, Bain, K, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-06 | | Release date: | 2008-09-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Methyltransferase Involved in Cell Division from Thermoplasma Volcanicum
To be Published
|
|
6HOW
 
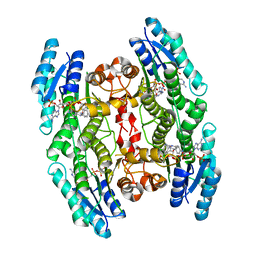 | | Trypanosoma brucei PTR1 in complex with the triazine inhibitor 2a (F219). | | Descriptor: | (2~{R})-1-(3,4-dichlorophenyl)-2-(4-nitrophenyl)-2~{H}-1,3,5-triazine-4,6-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Development of Cycloguanil Derivatives asTrypanosoma bruceiPteridine-Reductase-1 Inhibitors.
Acs Infect Dis., 5, 2019
|
|
3I5R
 
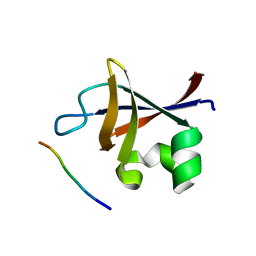 | | PI3K SH3 domain in complex with a peptide ligand | | Descriptor: | Peptide ligand, Phosphatidylinositol 3-kinase regulatory subunit alpha | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
3I5S
 
 | | Crystal structure of PI3K SH3 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION | | Authors: | Batra-Safferling, R, Granzin, J, Modder, S, Hoffmann, S, Willbold, D. | | Deposit date: | 2009-07-06 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.
Biol.Chem., 391, 2010
|
|
2GQ4
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ6
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ5
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3L8K
 
 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
3GO2
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium | | Descriptor: | MAGNESIUM ION, Putative L-alanine-DL-glutamate epimerase | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium.
To be Published
|
|
3GMG
 
 | | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv1825/MT1873 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis
To be Published
|
|
2LBU
 
 | | HADDOCK calculated model of Congo red bound to the HET-s amyloid | | Descriptor: | Small s protein, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | Authors: | Schutz, A.K, Soragni, A, Hornemann, S, Aguzzi, A, Ernst, M, Bockmann, A, Meier, B.H. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The Amyloid-Congo Red Interface at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 2011
|
|
2L6W
 
 | | PDGFR beta-TM | | Descriptor: | Beta-type platelet-derived growth factor receptor | | Authors: | Muhle-Goll, C, Hoffmann, S, Ulrich, A.S. | | Deposit date: | 2010-11-29 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic matching controls the tilt and stability of the dimeric platelet-derived growth factor receptor (PDGFR) beta transmembrane segment.
J.Biol.Chem., 287, 2012
|
|
2LT3
 
 | | Solution NMR structure of the C-terminal domain of CdnL from Myxococcus xanthus | | Descriptor: | Transcriptional regulator, CarD family | | Authors: | Mirassou, Y, Garcia-Moreno, D, Padmanabhan, S, Jimenez, M.A. | | Deposit date: | 2012-05-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into RNA Polymerase Recognition and Essential Function of Myxococcus xanthus CdnL.
Plos One, 9, 2014
|
|
