8W6G
 
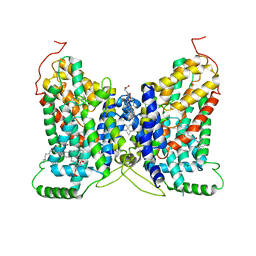 | | NaDC1 with inhibitor ACA | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-[[(~{E})-3-(4-pentylphenyl)prop-2-enoyl]amino]benzoic acid, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8W6T
 
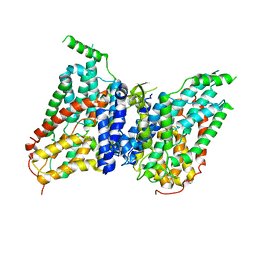 | | NaS1 in IN/OUT state | | Descriptor: | Solute carrier family 13 member 1 | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
8J5U
 
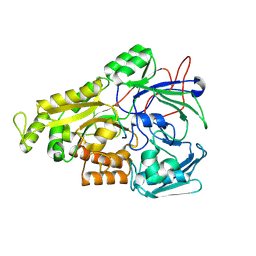 | | Crystal structure of Mycobacterium tuberculosis OppA complexed with an endogenous oligopeptide | | Descriptor: | Endogenous oligopeptide, Uncharacterized protein Rv1280c | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5S
 
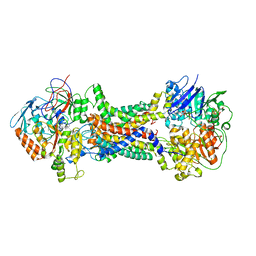 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-catalytic intermediate state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5R
 
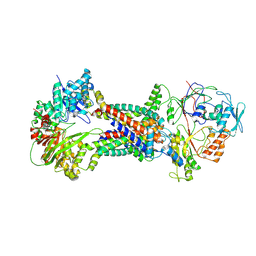 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the resting state | | Descriptor: | IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, Putative peptide transport permease protein Rv1283c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5T
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the catalytic intermediate state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5Q
 
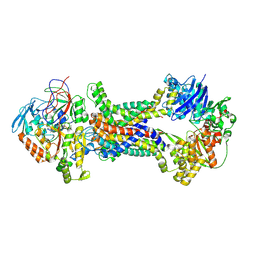 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-translocation state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8HPM
 
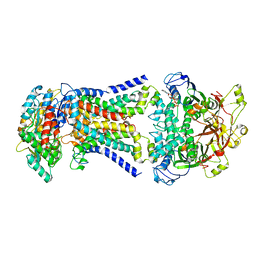 | | LpqY-SugABC in state 2 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
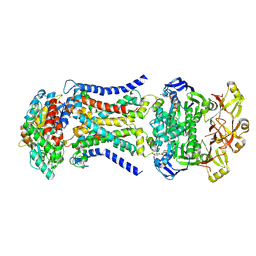 | | LpqY-SugABC in state 3 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
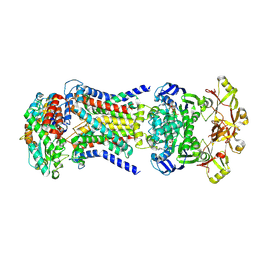 | | LpqY-SugABC in state 5 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPL
 
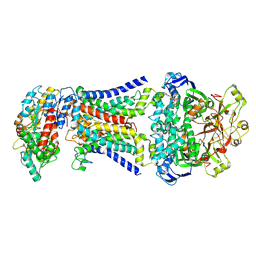 | | LpqY-SugABC in state 1 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPR
 
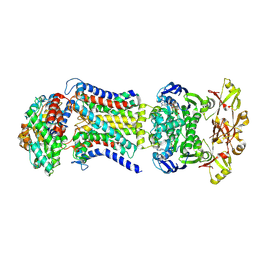 | | LpqY-SugABC in state 4 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
6KGT
 
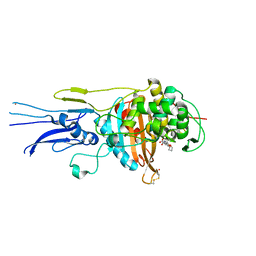 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
4WQO
 
 | | Structure of VHL-EloB-EloC-Cul2 | | Descriptor: | Cullin-2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Nguyen, H.C, Xiong, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into Cullin-RING E3 Ubiquitin Ligase Recruitment: Structure of the VHL-EloBC-Cul2 Complex.
Structure, 23, 2015
|
|
6KGS
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
6KGW
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
5ZZ9
 
 | | Crystal structure of Homer2 EVH1/Drebrin PPXXF complex | | Descriptor: | Homer protein homolog 2, Peptide from Drebrin | | Authors: | Li, Z, Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2018-05-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homer Tetramer Promotes Actin Bundling Activity of Drebrin.
Structure, 27, 2019
|
|
4PZA
 
 | | The complex structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c with inorganic phosphate | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, PHOSPHATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-29 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
6KGU
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
4PZ9
 
 | | The native structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-03-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
6KGV
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis, complexed with amoxicillin | | Descriptor: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, COBALT (II) ION, Penicillin-binding protein PbpB | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
4QIH
 
 | | The structure of mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c complexes with VO3 | | Descriptor: | Glucosyl-3-phosphoglycerate phosphatase, VANADATE ION | | Authors: | Zhou, W.H, Zheng, Q.Q, Jiang, D.Q, Zhang, W, Zhang, Q.Q, Jin, J, Li, X, Yang, H.T, Shaw, N, Rao, Z. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase
J.Biol.Chem., 289, 2014
|
|
8IZE
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IXV
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A in complex with 2Cl-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
