1AY4
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE WITHOUT SUBSTRATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY5
 
 | | AROMATIC AMINO ACID AMINOTRANSFERASE COMPLEX WITH MALEATE | | Descriptor: | AROMATIC AMINO ACID AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okamoto, A, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 1997-11-14 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network.
J.Mol.Biol., 280, 1998
|
|
1AY8
 
 | |
8HHV
 
 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
7CJI
 
 | | Photosystem II structure in the S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
8X6R
 
 | | KRasG12C in complex with inhibitor | | Descriptor: | 1-[7-[6-ethenyl-8-ethoxy-7-(5-methyl-1~{H}-indazol-4-yl)-2-(1-methylpiperidin-4-yl)oxy-quinazolin-4-yl]-2,7-diazaspiro[3.5]nonan-2-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ASP6918, a KRAS G12C inhibitor: Synthesis and structure-activity relationships of 1-{2,7-diazaspiro[3.5]non-2-yl}prop-2-en-1-one derivatives as covalent inhibitors with good potency and oral activity for the treatment of solid tumors.
Bioorg.Med.Chem., 98, 2023
|
|
5XSZ
 
 | | Crystal structure of zebrafish lysophosphatidic acid receptor LPA6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lysophosphatidic acid receptor 6a,Endolysin,Lysophosphatidic acid receptor 6a | | Authors: | Taniguchi, R, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA6
Nature, 548, 2017
|
|
7XY8
 
 | | Crystal structure of antibody Fab fragment in complex with CD147(EMMPIRIN) | | Descriptor: | Isoform 2 of Basigin, heavy chain, light chain | | Authors: | Nakamura, K, Amano, M, Yoneda, K, Suzuki, M, Fukuchi, K. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel Antibody Exerts Antitumor Effect through Downregulation of CD147 and Activation of Multiple Stress Signals.
J Oncol, 2022, 2022
|
|
6UMW
 
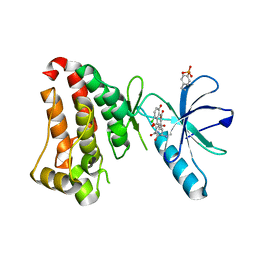 | | Crystal structure of hEphB1 bound with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2019-10-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2DQI
 
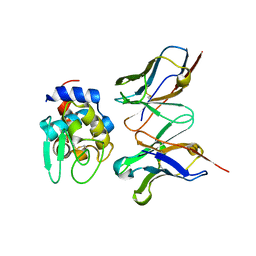 | | Crystal structure of hyhel-10 FV mutant (Ly50a) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-26 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2DQF
 
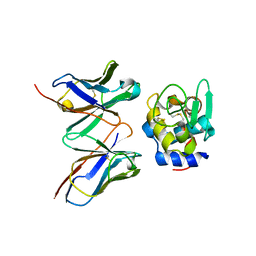 | | Crystal structure of hyhel-10 FV mutant (y33ay53a) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2DQG
 
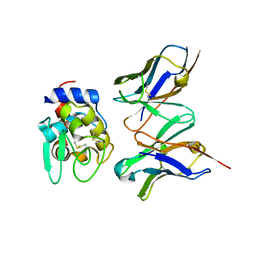 | | Crystal structure of hyhel-10 FV mutant (Hy53f) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2DQD
 
 | | Crystal structure of hyhel-10 FV mutant (Hy50f) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2DQC
 
 | | Crystal structure of hyhel-10 FV mutant(Hy33f) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
2DQH
 
 | | Crystal structure of hyhel-10 FV mutant (Hy58a) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-26 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
8I1C
 
 | |
8I1A
 
 | |
8I1I
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1D
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I18
 
 | |
8I1H
 
 | |
