1KXJ
 
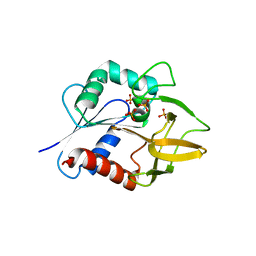 | | The Crystal Structure of Glutamine Amidotransferase from Thermotoga maritima | | Descriptor: | Amidotransferase hisH, PHOSPHATE ION | | Authors: | Korolev, S, Skarina, T, Evdokimova, E, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Thermotoga maritima
Proteins, 49, 2002
|
|
1KTN
 
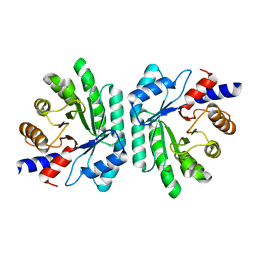 | | Structural Genomics, Protein EC1535 | | Descriptor: | 2-deoxyribose-5-phosphate aldolase | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Skarina, T, Evdokimova, E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-16 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.5A crystal structure of
2-deoxyribose-5-phosphate aldlase
To be Published
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1NC7
 
 | | Crystal Structure of Thermotoga maritima 1070 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-04 | | Release date: | 2003-07-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima Hypothetical protein TM1070
To be Published
|
|
1NJK
 
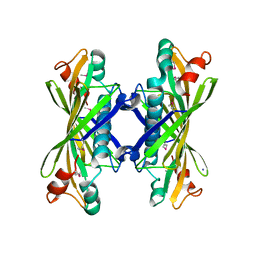 | | Crystal Structure of YbaW Probable Thioesterase from Escherichia coli | | Descriptor: | Hypothetical protein ybaW, IODIDE ION | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-31 | | Release date: | 2003-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Escherichia coli Hypothetical Protein YbaW
To be Published
|
|
2AP3
 
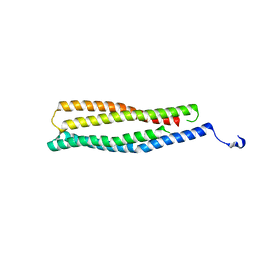 | | 1.6 A Crystal Structure of a Conserved Protein of Unknown Function from Staphylococcus aureus | | Descriptor: | conserved hypothetical protein | | Authors: | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a conserved hypothetical protein from Staphylococcus aureus MW2
To be Published
|
|
1XEB
 
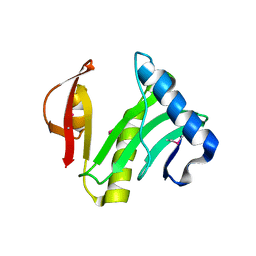 | | Crystal Structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA0115 | | Authors: | Bertero, M.G, Walker, J.R, Skarina, T, Gorodichtchenskaia, E, Joachimiak, A, Edwards, A.E, Savchenko, A, Strynadka, N, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of an Acyl-CoA N-acyltransferase from Pseudomonas aeruginosa
To be Published
|
|
1NI9
 
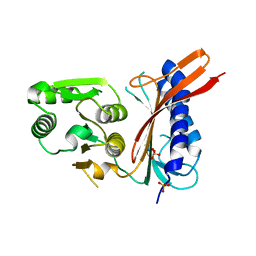 | | 2.0 A structure of glycerol metabolism protein from E. coli | | Descriptor: | Protein glpX, SULFATE ION | | Authors: | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-23 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
1ML8
 
 | | structural genomics | | Descriptor: | hypothetical protein (crp region) | | Authors: | Korolev, S, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-30 | | Release date: | 2003-04-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics
To be Published
|
|
1R8K
 
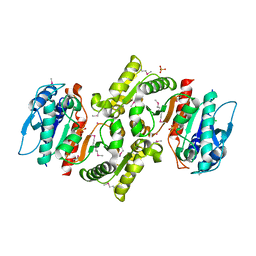 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
1NR9
 
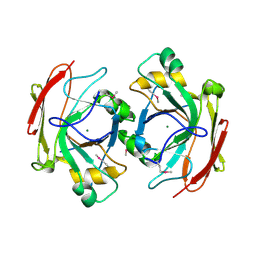 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | Descriptor: | MAGNESIUM ION, Protein YCGM | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
1UXO
 
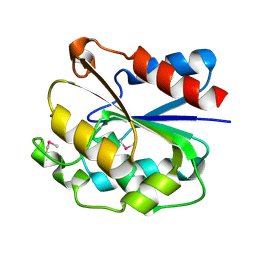 | | The crystal structure of the ydeN gene product from B. subtilis | | Descriptor: | Putative hydrolase YdeN | | Authors: | Janda, I.K, Devedjiev, Y, Cooper, D.R, Chruszcz, M, Derewenda, U, Gabrys, A, Minor, W, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-27 | | Release date: | 2004-05-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
Acta Crystallogr. D Biol. Crystallogr., 60, 2004
|
|
1NQK
 
 | | Structural Genomics, Crystal structure of Alkanesulfonate monooxygenase | | Descriptor: | Alkanesulfonate monooxygenase | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the protein
Alkanesulfonate monooxygenase from E. Coli
To be Published
|
|
1X87
 
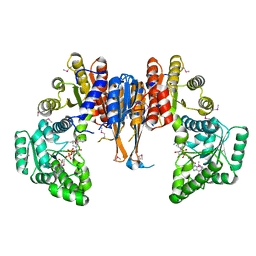 | | 2.4A X-ray structure of Urocanase protein complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Urocanase protein | | Authors: | Tereshko, V, Zaborske, J, Gilbreth, R, Dementieva, I, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A X-ray structure of Urocanase protein complexed with NAD
To be Published
|
|
1U14
 
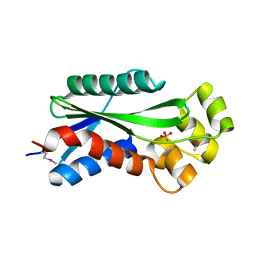 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
6D0P
 
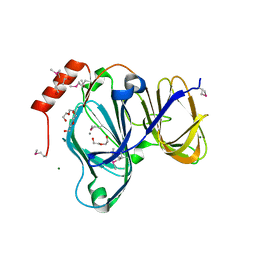 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
4EDH
 
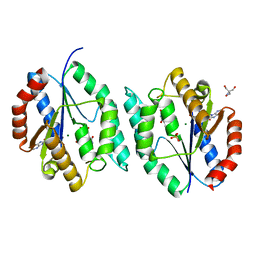 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg.
To be Published
|
|
4ESH
 
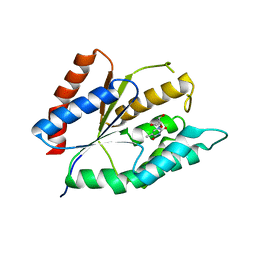 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine. | | Descriptor: | THYMIDINE, Thymidylate kinase | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with deoxythymidine.
To be Published
|
|
1KR4
 
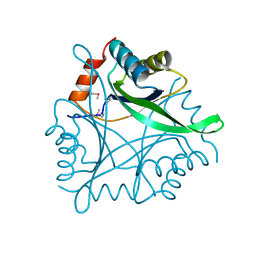 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
6D0G
 
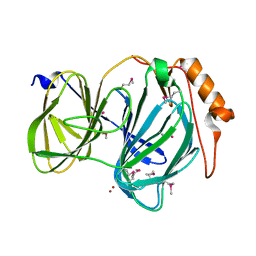 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
6DB1
 
 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | Descriptor: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6D72
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions.
To Be Published
|
|
3SR3
 
 | | Crystal structure of the w180a mutant of microcin immunity protein mccf from Bacillus anthracis shows the active site loop in the open conformation. | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-06 | | Release date: | 2011-08-10 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
6DGI
 
 | |
