7G9W
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z444860982 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, N-[(3,5-dimethyl-1H-pyrazol-4-yl)methyl]cyclohexanamine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GA1
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with POB0128 | | Descriptor: | (1S,2R)-2-(thiophen-3-yl)cyclopentane-1-carboxylic acid, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G9P
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1619958679 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-fluoro-N-(2-hydroxy-6-methylphenyl)pyridine-4-carboxamide, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GA0
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z2048325751 | | Descriptor: | (3R)-3-[1-methyl-4-(trifluoromethyl)-1H-imidazol-2-yl]piperidine, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G9N
 
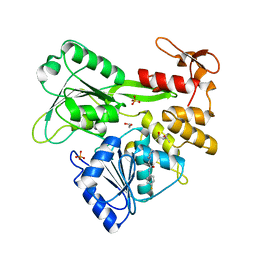 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z57821475 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, N-(4-fluorophenyl)-2-(piperazin-1-yl)acetamide, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.422 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GA3
 
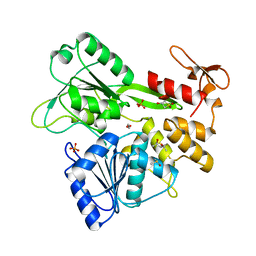 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z3241250482 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1-bis(oxidanylidene)thietan-3-ol, 1,2-ETHANEDIOL, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G9X
 
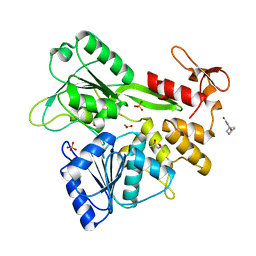 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z905065822 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5R)-1,3,7-triazaspiro[4.5]decane-2,4-dione, 1,2-ETHANEDIOL, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GA4
 
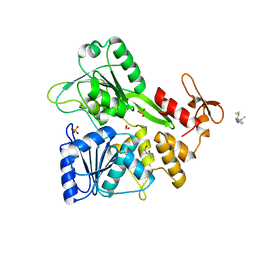 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56823075 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GA5
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z119989094 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(piperazin-1-yl)-1,3-benzothiazole, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GA7
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z85933875 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-[(3-fluoro-4-methoxyphenyl)methyl]piperazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7G9L
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z111782404 | | Descriptor: | (2S)-2-[([1,1'-biphenyl]-4-yl)oxy]propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5TGG
 
 | |
2LSC
 
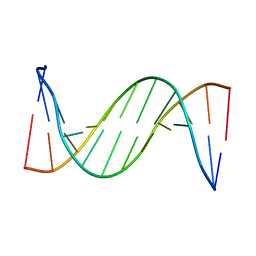 | | Solution structure of 2'F-ANA and ANA self-complementary duplex | | Descriptor: | DNA (5'-D(*(CFL)P*(GFL)P*(CFL)P*(GFL)P*(A5O)P*(A5O)P*(UAR)P*(UAR)P*(CFL)P*(GFL)P*(CFL)P*(GFL))-3') | | Authors: | Martin-Pintado, N, Yahyaee, M, Campos, R, Noronha, A, Wilds, C, Damha, M, Gonzalez, C. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of double helical arabino nucleic acids (ANA and 2'F-ANA): effect of arabinoses in duplex-hairpin interconversion.
Nucleic Acids Res., 40, 2012
|
|
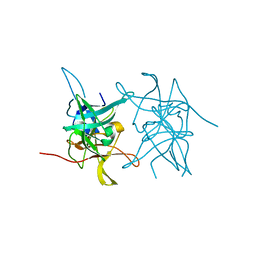
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of EV D68 3C protease - to be published
To Be Published
|
|
8CO8
 
 | | Structure of West Nile Virus NS2B-NS3 protease | | Descriptor: | Serine protease subunit NS2B,Serine protease/Helicase NS3 | | Authors: | Fairhead, M, Godoy, A.S, Koekemoer, L, Balcomb, B.H, Lithgo, R.M, Aschenbrenner, J.C, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-27 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of West Nile Virus NS2B-NS3 protease - to be published
To Be Published
|
|
6BJO
 
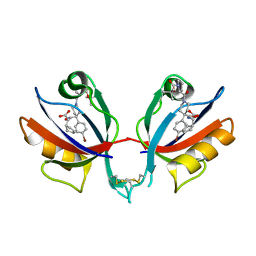 | | PICK1 PDZ domain in complex with the small molecule inhibitor BIO124. | | Descriptor: | (2S)-({4-(4-bromophenyl)-1-[1-(tert-butoxycarbonyl)-L-prolyl]piperidine-4-carbonyl}amino)(cyclopentyl)acetic acid, PRKCA-binding protein | | Authors: | Marcotte, D.J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-10 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Lock and chop: A novel method for the generation of a PICK1 PDZ domain and piperidine-based inhibitor co-crystal structure.
Protein Sci., 27, 2018
|
|
6BJN
 
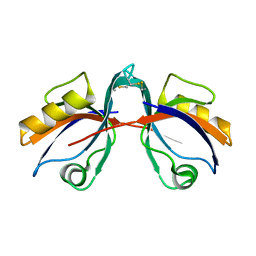 | |
7GB3
 
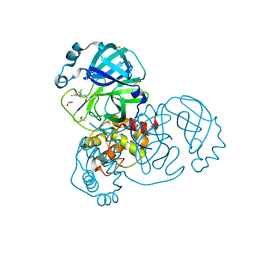 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAV-CRI-3edb475e-6 (Mpro-x10172) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1S)-1-(3-chloro-5-fluorophenyl)ethyl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBH
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-49816e9b-2 (Mpro-x10334) | | Descriptor: | 2-(3-chlorophenyl)-N-(2,4-dimethylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-8 (Mpro-x10423) | | Descriptor: | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCE
 
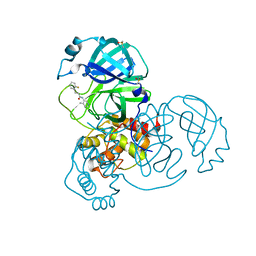 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BEN-DND-93268d01-7 (Mpro-x10598) | | Descriptor: | 3-(2-fluorophenyl)-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GCV
 
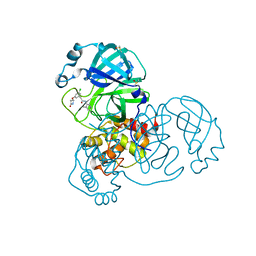 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-2eddb1ff-7 (Mpro-x10789) | | Descriptor: | 2-(3-chloro-5-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD2
 
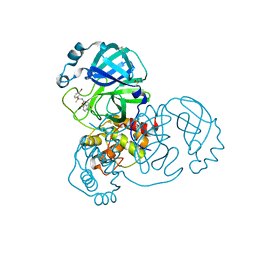 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-3 (Mpro-x10862) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-1-(3-bromophenyl)-2-methoxyethyl]-2-[(3S)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDD
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ADA-UCB-6c2cb422-1 (Mpro-x10959) | | Descriptor: | 2-(3-chlorophenyl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDI
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-49816e9b-1 (Mpro-x11011) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-2-(4-methylpyridin-3-yl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
