5TGS
 
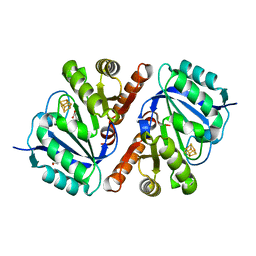 | | Crystal Structure of QueE from Bacillus subtilis with methionine bound | | Descriptor: | 7-carboxy-7-deazaguanine synthase, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
5TK6
 
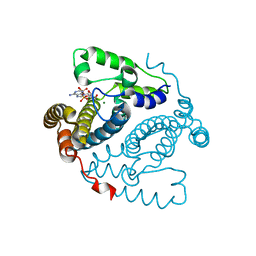 | |
5TH5
 
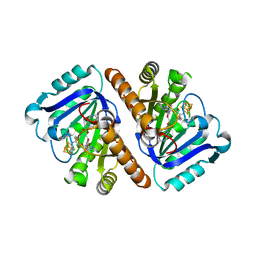 | | Crystal Structure of QueE from Bacillus subtilis with 6-carboxypterin-5'-deoxyadenosyl ester bound | | Descriptor: | 5'-O-(2-amino-4-oxo-1,4-dihydropteridine-6-carbonyl)adenosine, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
5TK8
 
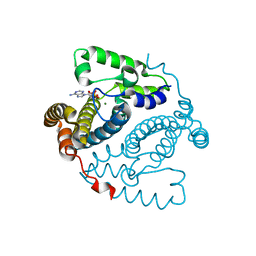 | |
5TK9
 
 | | Structure of the HD-domain phosphohydrolase OxsA with Oxetanocin-A bound | | Descriptor: | MAGNESIUM ION, OxsA protein, [(2S,3R,4R)-4-(6-amino-9H-purin-9-yl)oxetane-2,3-diyl]dimethanol | | Authors: | Bridwell-Rabb, J, Drennan, C.L. | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5TRB
 
 | | Crystal structure of the RNF20 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1A, ZINC ION | | Authors: | Foglizzo, M, Middleton, A.J, Day, C.L. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B.
J.Mol.Biol., 428, 2016
|
|
1TSI
 
 | |
5TU7
 
 | |
5TU9
 
 | |
5UFX
 
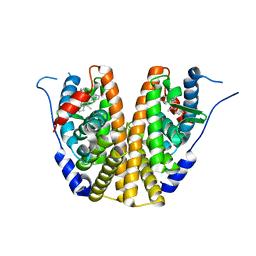 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1074 | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-4-methyl-2-(4-{2-[(3R)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallagher, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5503 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
5UFW
 
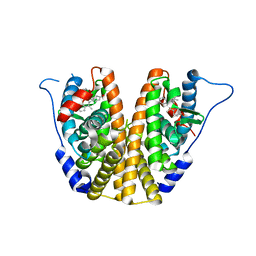 | | Estrogen Receptor Alpha Ligand Binding Domain in Complex with OP1154 | | Descriptor: | (2S)-3-(4-hydroxyphenyl)-4-methyl-2-(4-{2-[(3S)-3-methylpyrrolidin-1-yl]ethoxy}phenyl)-2H-1-benzopyran-7-ol, Estrogen receptor | | Authors: | Fanning, S.W, Hodges-Gallagher, L, Myles, D.C, Sun, R, Fowler, C.E, Green, B.D, Harmon, C.L, Greene, G.L, Kushner, P.J. | | Deposit date: | 2017-01-06 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Specific stereochemistry of OP-1074 disrupts estrogen receptor alpha helix 12 and confers pure antiestrogenic activity.
Nat Commun, 9, 2018
|
|
5UL2
 
 | |
5ULH
 
 | |
5UL3
 
 | | Structure of Cobalamin-dependent S-adenosylmethionine radical enzyme OxsB with aqua-cobalamin bound | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Bridwell-Rabb, J, Drennan, C.L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis.
Nature, 544, 2017
|
|
5V3T
 
 | | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis | | Descriptor: | CYANIDE ION, Globin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Varnado, C.L, SoRelle, E, Soman, J, Olson, J.S. | | Deposit date: | 2017-03-08 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis
To Be Published
|
|
5V3V
 
 | | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis: Tyr26Ala Mutant | | Descriptor: | CYANIDE ION, Globin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Varnado, C.L, SoRelle, E, Soman, J, Olson, J.S. | | Deposit date: | 2017-03-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of the Group II Truncated Hemoglobin from Bacillus Anthracis
To Be Published
|
|
5UL4
 
 | |
5ULK
 
 | |
5T81
 
 | | Rhombohedral crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB, GLYCEROL | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T8Y
 
 | | Structure of epoxyqueuosine reductase from Bacillus subtilis with the Asp134 catalytic loop swung out of the active site. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Maiocco, S.J, Elliott, S.J, Bandarian, V, Drennan, C.L. | | Deposit date: | 2016-09-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5TK7
 
 | |
5TKA
 
 | |
5UIC
 
 | |
5T5X
 
 | |
5T78
 
 | | Crystal structure of therapeutic mAB AR20.5 in complex with MUC1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Fab Fragment - AR20.5 - Heavy chain, Fab fragment AR20.5 - Light Chain, ... | | Authors: | Brooks, C.L, Movahedin, M. | | Deposit date: | 2016-09-02 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen.
Glycobiology, 27, 2017
|
|
