1B5W
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
5WTP
 
 | | Crystal structure of the C-terminal domain of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | OmpA family protein, SULFATE ION | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
5WTL
 
 | | Crystal structure of the periplasmic portion of outer membrane protein A (OmpA) from Capnocytophaga gingivalis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, OmpA family protein, ... | | Authors: | Dai, S, Tan, K, Ye, S, Zhang, R. | | Deposit date: | 2016-12-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability.
Cell Calcium, 66, 2017
|
|
1B7R
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
1B5X
 
 | | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and x-ray analysis of six ser->ala mutants | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B5Y
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
1B7O
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
3V75
 
 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
3UZR
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside phosphotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form
TO BE PUBLISHED
|
|
1C46
 
 | |
1C43
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (HUMAN LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C45
 
 | | MUTANT HUMAN LYSOZYME WITH FOREIGN N-TERMINAL RESIDUES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 1999-08-03 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of foreign N-terminal residues on the conformational stability of human lysozyme.
Eur.J.Biochem., 266, 1999
|
|
1C7P
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME WITH FOUR EXTRA RESIDUES (EAEA) AT THE N-TERMINAL | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Goda, S, Takano, K, Yamagata, Y, Katakura, Y, Yutani, K. | | Deposit date: | 2000-02-29 | | Release date: | 2000-04-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effect of extra N-terminal residues on the stability and folding of human lysozyme expressed in Pichia pastoris.
Protein Eng., 13, 2000
|
|
1X12
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192D) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1X10
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192A) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
3IXZ
 
 | | Pig gastric H+/K+-ATPase complexed with aluminium fluoride | | Descriptor: | Potassium-transporting ATPase alpha, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Tani, K, Nishizawa, T, Fujiyoshi, Y. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.5 Å) | | Cite: | Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle
Embo J., 28, 2009
|
|
1Z8W
 
 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192I) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8X
 
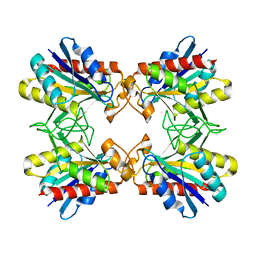 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192V) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1Z8T
 
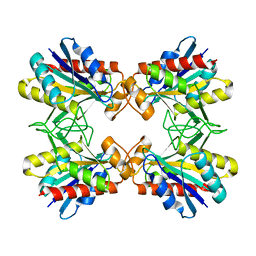 | | Structure of Mutant Pyrrolidone Carboxyl Peptidase (E192Q) from a Hyperthermophile, Pyrococcus furiosus | | Descriptor: | Pyrrolidone-carboxylate peptidase | | Authors: | Kaushik, J.K, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 2005-03-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Completely buried, non-ion-paired glutamic acid contributes favorably to the conformational stability of pyrrolidone carboxyl peptidases from hyperthermophiles.
Biochemistry, 45, 2006
|
|
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1TAB
 
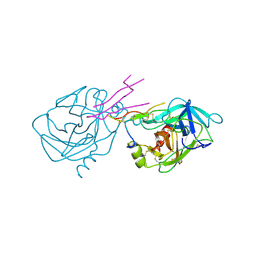 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | Descriptor: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | Authors: | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | Deposit date: | 1990-10-15 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
1CKC
 
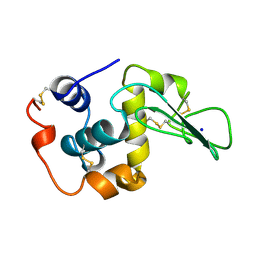 | | T43A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKG
 
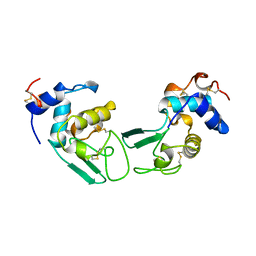 | | T52V MUTANT HUMAN LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CJ9
 
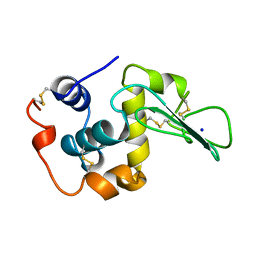 | | T40V MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
1CKF
 
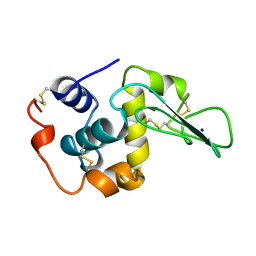 | | T52A MUTANT HUMAN LYSOZYME | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Funahashi, J, Yutani, K. | | Deposit date: | 1999-04-22 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,).
Biochemistry, 38, 1999
|
|
