7W23
 
 | | Structure of the M. tuberculosis HtrA S363A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W24
 
 | | Structure of the M. tuberculosis HtrA N383A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W25
 
 | | Structure of the M. tuberculosis HtrA S413A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VZ0
 
 | | Structure of the M. tuberculosis HtrA S407A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W21
 
 | | Structure of the M. tuberculosis HtrA N269A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7VYZ
 
 | | Structure of the M. tuberculosis HtrA S367A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7W4R
 
 | |
7W4W
 
 | |
7W4T
 
 | |
7W4U
 
 | |
7W4S
 
 | |
7W4V
 
 | |
3GI3
 
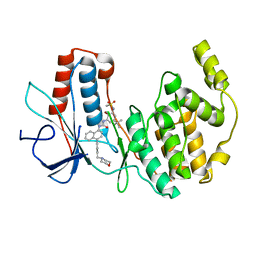 | | Crystal structure of a N-Phenyl-N'-Naphthylurea analog in complex with p38 MAP kinase | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-tert-butyl-2-methoxy-3-[({4-[6-(morpholin-4-ylmethyl)pyridin-3-yl]naphthalen-1-yl}carbamoyl)amino]phenyl}methanesulfonamide | | Authors: | Qian, K.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and characterization of the N-phenyl-N'-naphthylurea class of p38 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2LNA
 
 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
2L6U
 
 | | Solution NMR Structure of Med25(391-543) Comprising the Activator-Interacting Domain (ACID) of Human Mediator Subuniti 25. Northeast Structural Genomics Consortium Target HR6188A | | Descriptor: | Mediator complex subunit MED25 | | Authors: | Eletsky, A, Ryuechan, W.T, Sukumaran, D.K, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25.
J.Struct.Funct.Genom., 12, 2011
|
|
6Q7P
 
 | | Crystal structure of OE1.2 | | Descriptor: | 1,2-ETHANEDIOL, 1-PHENYLETHANONE, MAGNESIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7R
 
 | |
6Q7O
 
 | | Crystal structure of OE1 | | Descriptor: | CALCIUM ION, OE1 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7N
 
 | |
6QIY
 
 | | CI-2, conformation 1 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6Q7Q
 
 | | Crystal structure of OE1.3 | | Descriptor: | OE1.3 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6QIZ
 
 | | CI-2, conformation 2 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
2L0B
 
 | | Solution NMR structure of zinc finger domain of E3 ubiquitin-protein ligase praja-1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR4710B | | Descriptor: | E3 ubiquitin-protein ligase Praja-1, ZINC ION | | Authors: | Liu, G, Tong, S, Hamilton, K, Ciccosanti, C, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of zinc finger domain of E3 ubiquitin-protein ligase protein praja-1 from Homo sapiens, northeast structural genomics consortium (NESG) target HR4710B
To be Published
|
|
