7FDI
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
3ORF
 
 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | Descriptor: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
6FR2
 
 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
5JX8
 
 | |
5JX3
 
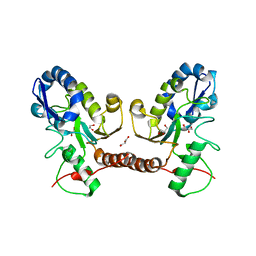 | | Wild type D4 in orthorhombic space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2016-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
5JX0
 
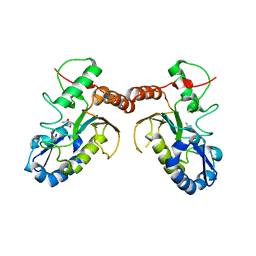 | | Temperature sensitive D4 mutant L110F | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2016-05-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Poxvirus uracil-DNA glycosylase-An unusual member of the family I uracil-DNA glycosylases.
Protein Sci., 25, 2016
|
|
3QNA
 
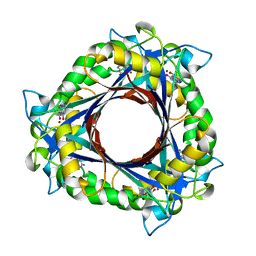 | | Crystal structure of a 6-pyruvoyltetrahydropterin synthase homologue from Esherichia coli complexed sepiapterin | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, BIOPTERIN, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Lee, K.H. | | Deposit date: | 2011-02-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a novel activity of bacterial 6-pyruvoyltetrahydropterin synthase homologues distinct from mammalian 6-pyruvoyltetrahydropterin synthase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QN9
 
 | |
3QN0
 
 | | Structure of 6-pyruvoyltetrahydropterin synthase | | Descriptor: | 6-carboxy-5,6,7,8-tetrahydropterin synthase, ZINC ION | | Authors: | Seo, K.H, Zhuang, N.N, Lee, K.H. | | Deposit date: | 2011-02-07 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis of a novel activity of bacterial 6-pyruvoyltetrahydropterin synthase homologues distinct from mammalian 6-pyruvoyltetrahydropterin synthase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5Z7I
 
 | | Caulobacter crescentus GcrA DNA-binding domain(DBD)in complex with unmethylated dsDNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*AP*TP*TP*CP*GP*C*)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5ZX3
 
 | |
5ZX2
 
 | |
5YIU
 
 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) | | Descriptor: | Cell cycle regulatory protein GcrA | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIW
 
 | | Caulobacter crescentus GcrA DNA-binding domain (DBD) in complex with methylated dsDNA (crystal form 2) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, DNA (5'-D(*CP*CP*CP*TP*GP*(6MA)P*TP*TP*CP*GP*C)-3'), ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
5YIV
 
 | |
5YIX
 
 | | Caulobacter crescentus GcrA sigma-interacting domain (SID) in complex with domain 2 of sigma 70 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cell cycle regulatory protein GcrA, RNA polymerase sigma factor RpoD, ... | | Authors: | Wu, X, Zhang, Y. | | Deposit date: | 2017-10-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Nucleic Acids Res., 46, 2018
|
|
4LHM
 
 | | Thymidine phosphorylase from E.coli with 3'-azido-3'-deoxythymidine | | Descriptor: | 3'-azido-3'-deoxythymidine, GLYCEROL, SULFATE ION, ... | | Authors: | Timofeev, V.I, Abramchik, Y.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2013-07-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | 3'-Azidothymidine in the active site of Escherichia coli thymidine phosphorylase: the peculiarity of the binding on the basis of X-ray study.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7WZZ
 
 | |
7X00
 
 | |
7X1B
 
 | |
7X1C
 
 | |
