5A60
 
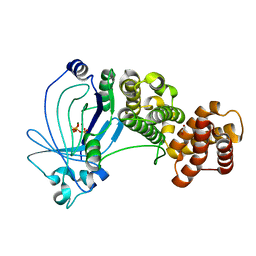 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two magnesium ions | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A68
 
 | |
2VXF
 
 | |
2WBR
 
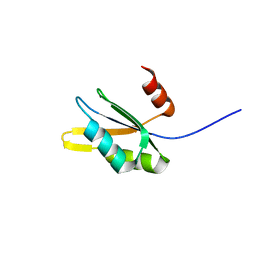 | | The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing | | Descriptor: | GW182 | | Authors: | Eulalio, A, Tritschler, F, Buettner, R, Weichenrieder, O, Izaurralde, E, Truffault, V. | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Rrm Domain in Gw182 Proteins Contributes to Mirna-Mediated Gene Silencing.
Nucleic Acids Res., 37, 2009
|
|
2VXE
 
 | |
5A67
 
 | |
4UZR
 
 | |
2VBU
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBV
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and FMN | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBT
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with CDP and PO4 | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Ammelburg, M, Djuranovic, S, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VBS
 
 | | Riboflavin kinase Mj0056 from Methanocaldococcus jannaschii in complex with PO4 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RIBOFLAVIN KINASE, ... | | Authors: | Hartmann, M.D, Djuranovic, S, Ammelburg, M, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2007-09-16 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Ctp-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
2VC8
 
 | |
2YKP
 
 | |
2YKQ
 
 | |
2YKO
 
 | |
6Y06
 
 | |
5IIG
 
 | |
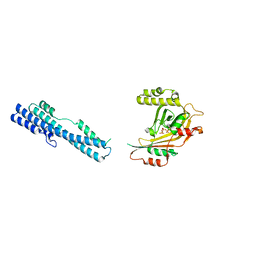
5IIT
 
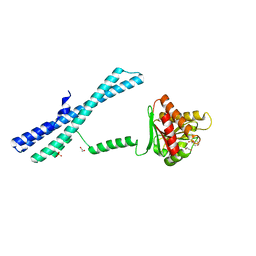 | |
5IJH
 
 | |
5IIQ
 
 | |
5IJJ
 
 | |
5IJP
 
 | |
2C0S
 
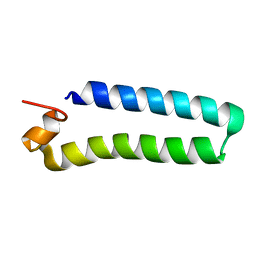 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2BZB
 
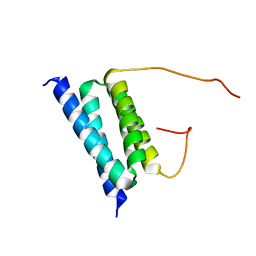 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
6Y07
 
 | |
