4U78
 
 | | Octameric RNA duplex soaked in copper(II)chloride | | Descriptor: | CALCIUM ION, COPPER (II) ION, RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3') | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-30 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
7QR3
 
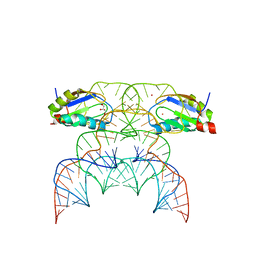 | | Chimpanzee CPEB3 HDV-like ribozyme | | Descriptor: | GLYCEROL, POTASSIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
7QR4
 
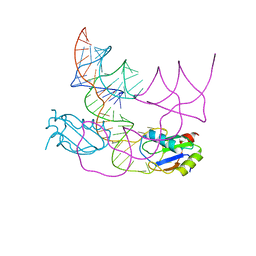 | | human CPEB3 HDV-like ribozyme | | Descriptor: | RNA CPEB3 ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
2M23
 
 | |
2K66
 
 | |
2KE8
 
 | | NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Johannsen, S, Duepre, N, Boehme, D, Mueller, J, Sigel, R.K.O. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA double helix with consecutive metal-mediated base pairs.
Nat.Chem., 2, 2010
|
|
2M24
 
 | |
2M1V
 
 | |
2M5U
 
 | |
