6HHM
 
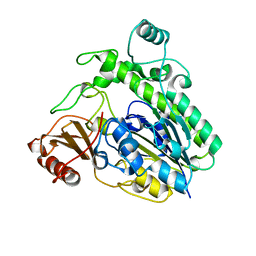 | | Crystal structure of the family S1_7 ulvan-specific sulfatase FA22070 from Formosa agariphila | | Descriptor: | Arylsulfatase, CALCIUM ION | | Authors: | Roret, T, Prechoux, A, Michel, G, Czjzek, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6HR5
 
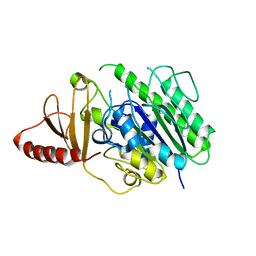 | | Structure of the S1_25 family sulfatase module of the rhamnosidase FA22250 from Formosa agariphila | | Descriptor: | Alpha-L-rhamnosidase/sulfatase (GH78), CALCIUM ION | | Authors: | Roret, T, Prechoux, A, Czjzek, M, Michel, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.912 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6G61
 
 | |
6G62
 
 | |
7BLY
 
 | | Structure of the chitin deacetylase AngCDA from Aspergillus niger | | Descriptor: | Aspergillus niger contig An12c0130, genomic contig, CHLORIDE ION, ... | | Authors: | Roret, T, Bonin, M, Hembach, L, Moerschbacher, B.M. | | Deposit date: | 2021-01-19 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | In silico and in vitro analysis of an Aspergillus niger chitin deacetylase to decipher its subsite sugar preferences.
J.Biol.Chem., 297, 2021
|
|
2N5F
 
 | |
8RZJ
 
 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor ADG-IF (3,6-anhydro-D-galacto-isofagomine). | | Descriptor: | (1~{R},5~{R},8~{S})-6-oxa-3-azabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZK
 
 | | The Michaelis complex of ZgGH129 D486N from Zobellia galactanivorans with neo-b/k-oligo-carrageenan tetrasaccharide (beta-kappa neo-oligo-carrageenan DP4). | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZH
 
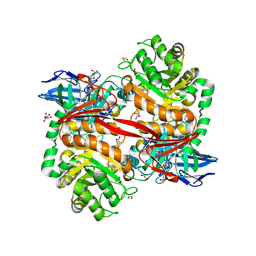 | | ZgGH129 from Zobellia galactanivorans in complex with the inhibitor AD-DGJ (3,6-anhydro-D-1-deoxygalactonojirimycin). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-6-oxa-2-azabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZI
 
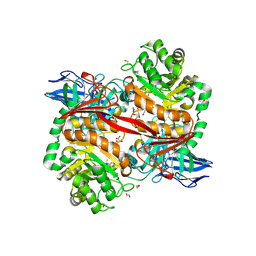 | | ZgGH129 from Zobellia galactanivorans soaked with 1,2-diF-ADG (3,6-Anhydro-2-deoxy-2-fluoro-a-D-galactopyranosyl fluoride) resulting in a trapped glycosyl-enzyme intermediate. | | Descriptor: | (1~{R},4~{S},5~{S},8~{S})-4-fluoranyl-2,6-dioxabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RZG
 
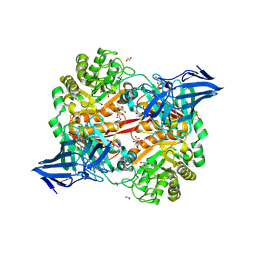 | | ZgGH129 from Zobellia galactanivorans soaked with the product of the reaction ADG (3,6-anhydro-D-galactose). | | Descriptor: | (1~{R},4~{S},5~{R},8~{S})-2,6-dioxabicyclo[3.2.1]octane-4,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5MYE
 
 | |
5N9U
 
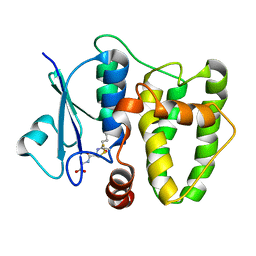 | |
7QNM
 
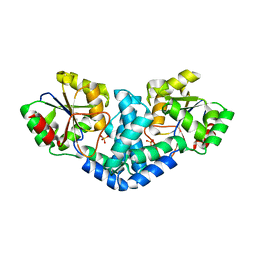 | | Crystallization and structural analyses of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION | | Authors: | Grigorian, E, Roret, T, Leblanc, C, Delage, L, Czjzek, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a l-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 32, 2023
|
|
4F0B
 
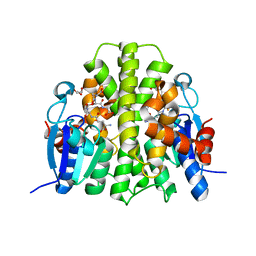 | |
7ASZ
 
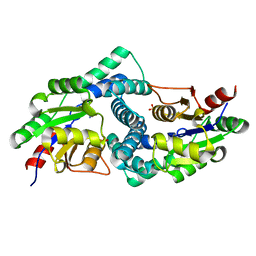 | | L-2-haloacid dehalogenase H190A mutant from Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION, THIOCYANATE ION | | Authors: | Grigorian, E, Roret, T, Czjzek, M, Leblanc, C, Delage, L. | | Deposit date: | 2020-10-28 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 2022
|
|
7ARP
 
 | | Native L-2-haloacid dehalogenase from Zobellia galactanivorans | | Descriptor: | (S)-2-haloacid dehalogenase, PHOSPHATE ION, THIOCYANATE ION | | Authors: | Grigorian, E, Roret, T, Czjzek, M, Leblanc, C, Delage, L. | | Deposit date: | 2020-10-26 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-ray structure and mechanism of ZgHAD, a L-2-haloacid dehalogenase from the marine Flavobacterium Zobellia galactanivorans.
Protein Sci., 2022
|
|
7BJT
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | Alginate lyase, family PL17, CALCIUM ION, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
7BM6
 
 | | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid, Alginate lyase, family PL17, ... | | Authors: | Czjzek, M, Roret, T, Jouanneau, D, Le Duff, N, Jeudy, A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT.
Glycobiology, 31, 2021
|
|
5IZ1
 
 | | Physcomitrella patens FBPase | | Descriptor: | fructose-1,6-bisphosphatase | | Authors: | Einsle, O, Guetle, D. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IZ3
 
 | | P. patens sedoheptulose-1,7-bisphosphatase | | Descriptor: | IMIDAZOLE, PHOSPHATE ION, Predicted protein, ... | | Authors: | Einsle, O, Guetle, D. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8CK0
 
 | |
8CK1
 
 | | Carin 1 bacteriophage tail, connector and tail fibers assembly | | Descriptor: | Connector Protein, Tail Nozzle, Tail fibers Dpo36 | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
8CJZ
 
 | | Carin1 bacteriophage mature capsid | | Descriptor: | Capsid Decoration Protein, Major Capsid Protein, Spike Base Protein | | Authors: | d'Acapito, A, Neumann, E, Schoehn, G. | | Deposit date: | 2023-02-14 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Study of the Cobetia marina Bacteriophage 1 (Carin-1) by Cryo-EM.
J.Virol., 97, 2023
|
|
6HPD
 
 | | The structure of a beta-glucuronidase from glycoside hydrolase family 2 | | Descriptor: | BROMIDE ION, Beta-galactosidase (GH2), MAGNESIUM ION | | Authors: | Robb, C.S, Gerlach, N, Reisky, L, Bornshoeru, U, Hehemann, J.H. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
