7VY7
 
 | |
7VXV
 
 | |
7VY4
 
 | |
5KSU
 
 | | Crystal structure of HLA-DQ2.5-CLIP1 at 2.73 resolution | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DQ alpha 1 chain, ... | | Authors: | Nguyen, T.-B, Jayaraman, P, Bergseng, E, Madhusudhan, M.S, Kim, C.-Y, Sollid, L.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Unraveling the structural basis for the unusually rich association of human leukocyte antigen DQ2.5 with class-II-associated invariant chain peptides.
J. Biol. Chem., 292, 2017
|
|
2IPK
 
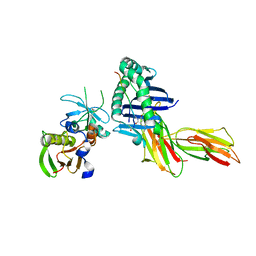 | | Crystal Structure of the MHC Class II Molecule HLA-DR1 in Complex with the Fluorogenic Peptide, AcPKXVKQNTLKLAT (X=3-[5-(dimethylamino)-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl]-L-alanine) and the Superantigen, SEC3 Variant 3B2 | | Descriptor: | Enterotoxin type C-3, HA related Fluorogenic Peptide, AcPKXVKQNTLKLAT (X=3-[5-(dimethylamino)-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl]-L-alanine), ... | | Authors: | Nguyen, T.T, Stern, L.J. | | Deposit date: | 2006-10-12 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fluorogenic probes for monitoring peptide binding to class II MHC proteins in living cells.
Nat.Chem.Biol., 3, 2007
|
|
3X1U
 
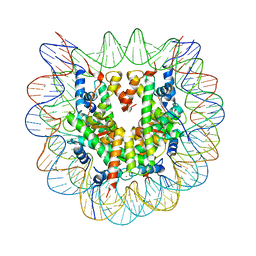 | |
3X1V
 
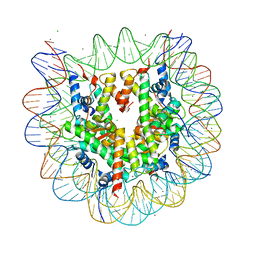 | |
3X1T
 
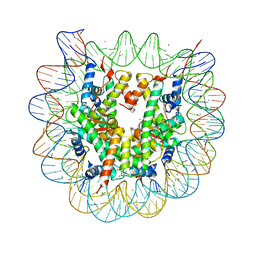 | |
3X1S
 
 | | Crystal structure of the nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Sivaraman, P, Kumarevel, T.S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural and functional analyses of nucleosome complexes with mouse histone variants TH2a and TH2b, involved in reprogramming
Biochem.Biophys.Res.Commun., 464, 2015
|
|
1DVS
 
 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN IN COMPLEX WITH RESVERATROL | | Descriptor: | RESVERATROL, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-22 | | Release date: | 2001-01-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
1DVQ
 
 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN | | Descriptor: | TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-22 | | Release date: | 2001-01-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
1DVT
 
 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN IN COMPLEX WITH FLURBIPROFEN | | Descriptor: | FLURBIPROFEN, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-22 | | Release date: | 2001-01-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
1DVY
 
 | | CRYSTAL STRUCTURE OF TRANSTHYRETIN IN COMPLEX WITH N-(M-TRIFLUOROMETHYLPHENYL) PHENOXAZINE-4,6-DICARBOXYLIC ACID | | Descriptor: | N-(M-TRIFLUOROMETHYLPHENYL) PHENOXAZINE-4,6-DICARBOXYLIC ACID, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-23 | | Release date: | 2001-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
1DVU
 
 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN IN COMPLEX WITH DIBENZOFURAN-4,6-DICARBOXYLIC ACID | | Descriptor: | DIBENZOFURAN-4,6-DICARBOXYLIC ACID, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-22 | | Release date: | 2001-01-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
1DVX
 
 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN IN COMPLEX WITH DICLOFENAC | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-23 | | Release date: | 2001-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
1DVZ
 
 | | CRYSTAL STRUCTURE OF HUMAN TRANSTHYRETIN IN COMPLEX WITH O-TRIFLUOROMETHYLPHENYL ANTHRANILIC ACID | | Descriptor: | O-TRIFLUOROMETHYLPHENYL ANTHRANILIC ACID, TRANSTHYRETIN | | Authors: | Klabunde, T, Petrassi, H.M, Oza, V.B, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2000-01-23 | | Release date: | 2001-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of potent human transthyretin amyloid disease inhibitors.
Nat.Struct.Biol., 7, 2000
|
|
6DKU
 
 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | Descriptor: | VP35 | | Authors: | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
3UDE
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1B | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UD5
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1A | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UDV
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1C | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
2G8A
 
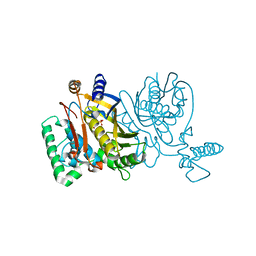 | | Lactobacillus casei Y261M in complex with substrate, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, thymidylate synthase | | Authors: | Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261
Biochemistry, 45, 2006
|
|
2G8X
 
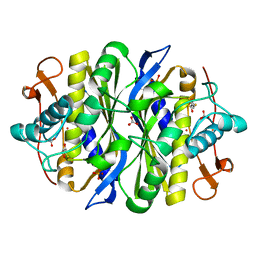 | | Escherichia coli Y209W apoprotein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Lee, T.T, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-03 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved dUMP-binding Tyr-261
TO BE PUBLISHED
|
|
2G89
 
 | |
2G86
 
 | |
2G8D
 
 | |
