1OH3
 
 | | E78R mutant of a carbohydrate binding module family 29 | | Descriptor: | NON-CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nurizzo, D, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ligand-Mediated Dimerization of a Carbohydrate -Binding Module Reveals a Novel Mechanism for Protein-Carbohydrate Recognition
J.Mol.Biol., 337, 2004
|
|
1GU3
 
 | | CBM4 structure and function | | Descriptor: | ENDOGLUCANASE C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1GUI
 
 | | CBM4 structure and function | | Descriptor: | CALCIUM ION, GLYCEROL, LAMINARINASE 16A, ... | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-27 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1GJQ
 
 | | Pseudomonas aeruginosa cd1 nitrite reductase reduced cyanide complex | | Descriptor: | CYANIDE ION, HEME C, HEME D, ... | | Authors: | Nurizzo, D, Brown, K, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanide Binding to Cd(1) Nitrite Reductase from Pseudomonas Aeruginosa: Role of the Active-Site His369 in Ligand Stabilization.
Biochem.Biophys.Res.Commun., 291, 2002
|
|
4QDN
 
 | | Crystal Structure of the endo-beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Flagellar protein FlgJ [peptidoglycan hydrolase], PHOSPHATE ION | | Authors: | Lipski, A, Nurizzo, D, Bourne, Y, Vincent, F. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of the beta-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family.
Glycobiology, 25, 2015
|
|
2H85
 
 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1QWD
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
1CNO
 
 | | STRUCTURE OF PSEUDOMONAS NAUTICA CYTOCHROME C552, BY MAD METHOD | | Descriptor: | CYTOCHROME C552, GLYCEROL, HEME C | | Authors: | Brown, K, Nurizzo, D, Cambillau, C. | | Deposit date: | 1998-08-03 | | Release date: | 1999-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association.
J.Mol.Biol., 289, 1999
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
2O84
 
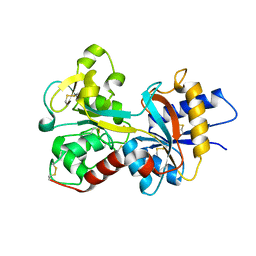 | | Crystal structure of K206E mutant of N-lobe human transferrin | | Descriptor: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | Authors: | Baker, H.M, Nurizzo, D, Mason, A.B, Baker, E.N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2O7U
 
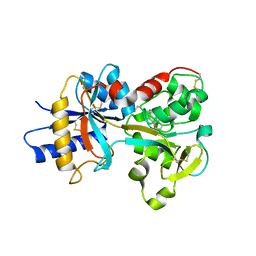 | | Crystal structure of K206E/K296E mutant of the N-terminal half molecule of human transferrin | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin | | Authors: | Baker, H.M, Nurizzo, D, Mason, A.B, Baker, E.N. | | Deposit date: | 2006-12-11 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2C8R
 
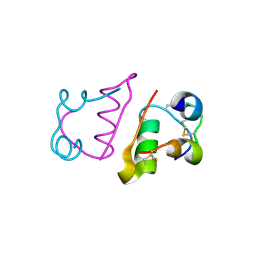 | | insuline(60sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C8O
 
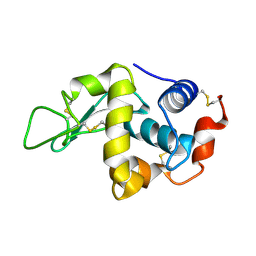 | | lysozyme (1sec) and UV lasr excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C8P
 
 | | lysozyme (60sec) and UV laser excited fluorescence | | Descriptor: | LYSOZYME C | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C8Q
 
 | | insuline(1sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2XFD
 
 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
1UWW
 
 | | X-ray crystal structure of a non-crystalline cellulose specific carbohydrate-binding module: CBM28. | | Descriptor: | CALCIUM ION, ENDOGLUCANASE | | Authors: | Jamal, S, Nurizzo, D, Boraston, A, Davies, G.J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Crystal Structure of a Non-Crystalline Cellulose-Specific Carbohydrate-Binding Module: Cbm28
J.Mol.Biol., 339, 2004
|
|
1WCU
 
 | | CBM29_1, A Family 29 Carbohydrate Binding Module from Piromyces equi | | Descriptor: | GLYCEROL, NON-CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davis, G.J, Gilbert, H.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
