6MEZ
 
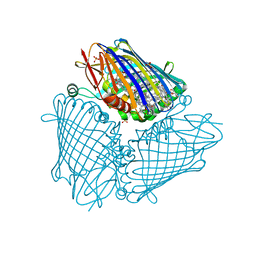 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | Authors: | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
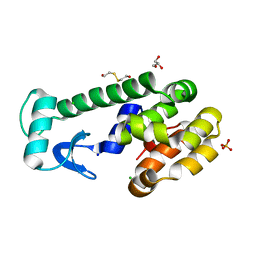 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
1GKT
 
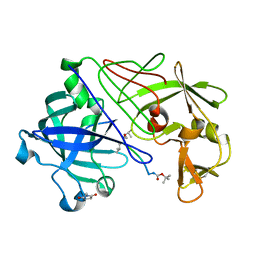 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
1OEX
 
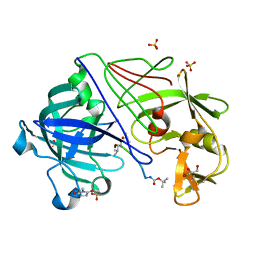 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEW
 
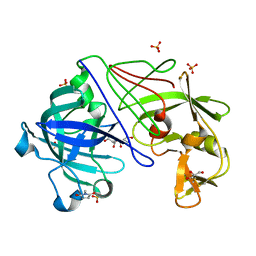 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
7ADQ
 
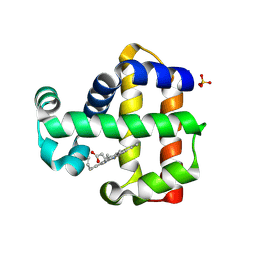 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
6N9H
 
 | | De novo designed homo-trimeric amantadine-binding protein | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, SODIUM ION, amantadine-binding protein | | Authors: | Park, J, Baker, D. | | Deposit date: | 2018-12-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.039 Å) | | Cite: | De novo design of a homo-trimeric amantadine-binding protein.
Elife, 8, 2019
|
|
7KFM
 
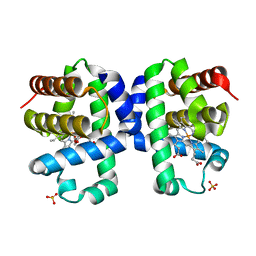 | | Room temperature oxyferrous Dehaloperoxidase B | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Dehaloperoxidase B, OXYGEN MOLECULE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
4K9F
 
 | | Neutron structure of Perdeuterated Rubredoxin refined against 1.75 resolution data collected on the new IMAGINE instrument at HFIR, ORNL | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-04-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | The IMAGINE instrument: first neutron protein structure and new capabilities for neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4LNC
 
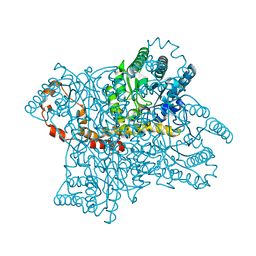 | | Neutron structure of the cyclic glucose bound Xylose Isomerase E186Q mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.19 Å) | | Cite: | Neutron structure of the cyclic glucose-bound xylose isomerase E186Q mutant.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7KCU
 
 | | Joint neutron/X-ray structure of Oxyferrous Dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Carey, L.M, Ghiladi, R.A, Meilleur, F, Myles, D. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
4JSD
 
 | |
4JSO
 
 | |
3KYU
 
 | |
3KYX
 
 | |
3KYV
 
 | |
3KYW
 
 | |
3KYY
 
 | |
1CWN
 
 | | CRYSTAL STRUCTURE OF PORCINE ALDEHYDE REDUCTASE HOLOENZYME | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O. | | Deposit date: | 1996-07-30 | | Release date: | 1998-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of porcine aldehyde reductase at 2.0 angstrom resolution: Modeling an inhibitor in the active site of the enzyme.
Protein Pept.Lett., 3, 1996
|
|
1AE4
 
 | |
4PFT
 
 | | Crystal structure of mannobiose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1223) from THERMOTOGA MARITIMA at 1.75 A resolution | | Descriptor: | ABC transporter substrate-binding protein, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
4PFW
 
 | | Crystal structure of mannohexaose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.2 A resolution | | Descriptor: | ABC transporter substrate-binding protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
4PFY
 
 | | Crystal structure of mannohexaose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1223) from THERMOTOGA MARITIMA at 1.5 A resolution | | Descriptor: | ABC transporter substrate-binding protein, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
4PFU
 
 | | Crystal structure of mannobiose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.05 A resolution | | Descriptor: | ABC transporter substrate-binding protein, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-04-30 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
