3CEX
 
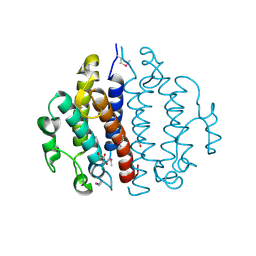 | | Crystal structure of the conserved protein of locus EF_3021 from Enterococcus faecalis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Cuff, M.E, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the conserved protein of locus EF_3021 from Enterococcus faecalis.
TO BE PUBLISHED
|
|
3FGX
 
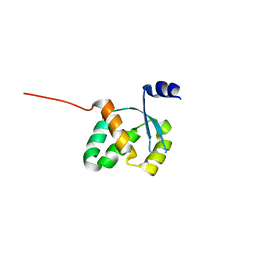 | | Structure of uncharacterised protein rbstp2171 from bacillus stearothermophilus | | Descriptor: | RBSTP2171 | | Authors: | Minasov, G, Filippova, E.V, Shuvalova, L, Moy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Uncharacterised Protein Rbstp2171 from Bacillus Stearothermophilus
To be Published
|
|
3FET
 
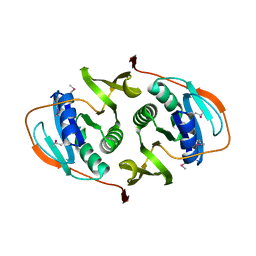 | | Crystal Structure of the Electron Transfer Flavoprotein Subunit Alpha related Protein Ta0212 from Thermoplasma acidophilum | | Descriptor: | Electron transfer flavoprotein subunit alpha related protein | | Authors: | Kim, Y, Tesar, C, Souvong, K, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Electron Transfer Flavoprotein Subunit Alpha related Protein Ta0212 from Thermoplasma acidophilum
To be Published
|
|
3DF8
 
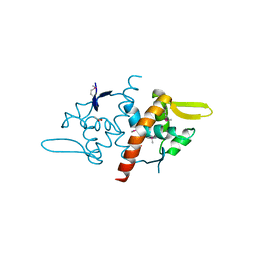 | |
3FTB
 
 | |
3BRJ
 
 | | Crystal structure of mannitol operon repressor (MtlR) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mannitol operon repressor | | Authors: | Tan, K, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
2EW2
 
 | | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis | | Descriptor: | 2-dehydropantoate 2-reductase, putative, MAGNESIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis
To be Published
|
|
3BIO
 
 | | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of oxidoreductase (Gfo/Idh/MocA family member) from Porphyromonas gingivalis W83.
To be Published
|
|
3BJO
 
 | |
3BED
 
 | |
3B48
 
 | |
3BP1
 
 | | Crystal structure of putative 7-cyano-7-deazaguanine reductase QueF from Vibrio cholerae O1 biovar eltor | | Descriptor: | GUANINE, MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-08 | | Last modified: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism.
J.Mol.Biol., 404, 2010
|
|
2IQT
 
 | | Crystal Structure of Fructose-Bisphosphate Aldolase, Class I from Porphyromonas gingivalis | | Descriptor: | Fructose-bisphosphate aldolase class 1 | | Authors: | Kim, Y, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-14 | | Release date: | 2006-11-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structure of Fructose-Bisphosphate Aldolase, Class I from Porphyromonas gingivalis
To be Published
|
|
2I6D
 
 | | The structure of a putative RNA methyltransferase of the TrmH family from Porphyromonas gingivalis. | | Descriptor: | ACETIC ACID, RNA methyltransferase, TrmH family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-28 | | Release date: | 2006-10-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of a putative RNA methyltransferase of the TrmH family from Porphyromonas gingivalis.
TO BE PUBLISHED
|
|
2I7H
 
 | | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitroreductase-like family protein, SULFATE ION | | Authors: | Kim, Y, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nitroreductase-like Family Protein from Bacillus cereus
To be Published
|
|
2IQQ
 
 | | The Crystal Structure of Iron, Sulfur-Dependent L-serine dehydratase from Legionella pneumophila subsp. pneumophila | | Descriptor: | Iron, Sulfur-Dependent L-serine dehydratase, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos, C, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-14 | | Release date: | 2006-11-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Iron, Sulfur-Dependent L-serine dehydratase from Legionella pneumophila subsp. pneumophila
To be Published
|
|
2IAF
 
 | |
2GWN
 
 | | The structure of putative dihydroorotase from Porphyromonas gingivalis. | | Descriptor: | BETA-MERCAPTOETHANOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Borovilos, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of putative dihydroorotase from Porphyromonas gingivalis.
To be Published
|
|
2H5N
 
 | |
2GUK
 
 | |
2GU3
 
 | | YpmB protein from Bacillus subtilis | | Descriptor: | NICKEL (II) ION, YpmB protein | | Authors: | Osipiuk, J, Maltseva, N, Dementieva, I, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray crystal structure of YpmB protein from Bacillus subtilis.
To be Published
|
|
2I6X
 
 | | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. (CASP Target)
To be Published
|
|
2I00
 
 | | Crystal structure of acetyltransferase (GNAT family) from Enterococcus faecalis | | Descriptor: | Acetyltransferase, GNAT family | | Authors: | Zhang, R, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-09 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the acetyltransferase (GNAT family) from Enterococcus faecalis
To be Published, 2006
|
|
1PZX
 
 | | Hypothetical protein APC36103 from Bacillus stearothermophilus: a lipid binding protein | | Descriptor: | Hypothetical protein APC36103, PALMITIC ACID | | Authors: | Zhang, R, Osipiuk, J, Zhou, M, Alkire, R, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-07-14 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipid binding protein APC36103 from Bacillus Stearothermophilus
To be Published
|
|
1Q77
 
 | | X-ray crystal structure of putative Universal Stress Protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_178, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-16 | | Release date: | 2003-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural homolog of Universal Stress Protein from Aquifex aeolicus
To be Published
|
|
