6BI6
 
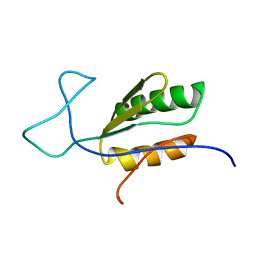 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
7TO8
 
 | |
7TOA
 
 | |
2HVA
 
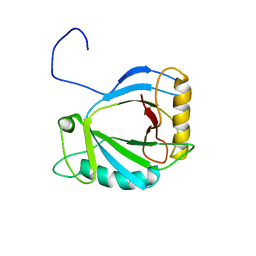 | | Solution Structure of the haem-binding protein p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Gell, D.A, Mackay, J.P, Westman, B.J, Liew, C.K, Gorman, D. | | Deposit date: | 2006-07-28 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Novel Haem-binding Interface in the 22 kDa Haem-binding Protein p22HBP.
J.Mol.Biol., 362, 2006
|
|
4F2J
 
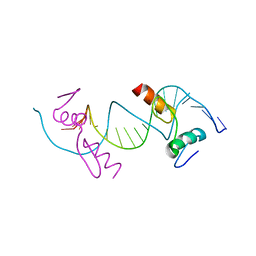 | | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | | Descriptor: | 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', ZINC ION, Zinc finger protein 217 | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2012-05-08 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217
J.Biol.Chem., 288, 2013
|
|
4PC0
 
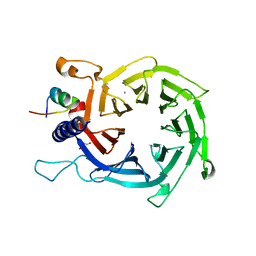 | | Structure of the human RbAp48-MTA1(670-711) complex | | Descriptor: | CALCIUM ION, GLYCEROL, Histone-binding protein RBBP4, ... | | Authors: | Alqarni, S.S.M, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4PBZ
 
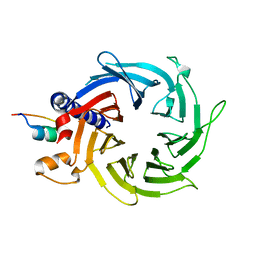 | | Structure of the human RbAp48-MTA1(670-695) complex | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Pei, X.Y, Watson, A.A, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4PBY
 
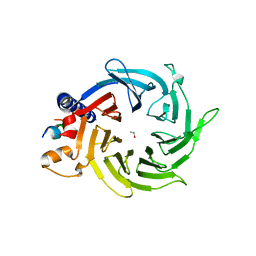 | | Structure of the human RbAp48-MTA1(656-686) complex | | Descriptor: | Histone-binding protein RBBP4, ISOPROPYL ALCOHOL, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Lejon, S, Alqarni, S.S.M, Silva, A.P.G, Watson, A.A, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
1J2O
 
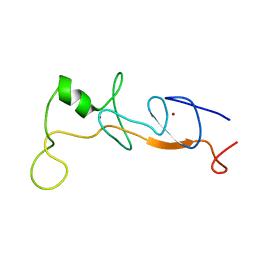 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | Descriptor: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
2XU7
 
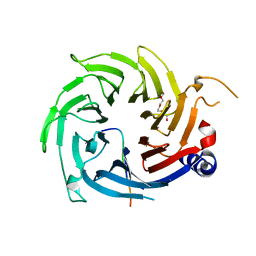 | | Structural basis for RbAp48 binding to FOG-1 | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, TETRAETHYLENE GLYCOL, ZINC FINGER PROTEIN ZFPM1 | | Authors: | Lejon, S, Thong, S.Y, Murthy, A, Blobel, G.A, Mackay, J.P, Murzina, N.V, Laue, E.D. | | Deposit date: | 2010-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights Into Association of the Nurd Complex with Fog-1 from the Crystal Structure of an Rbap48-Fog- 1 Complex.
J.Biol.Chem., 286, 2011
|
|
1MM2
 
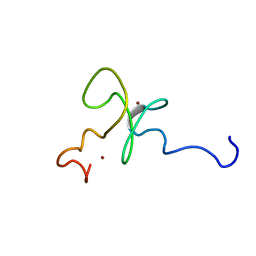 | | Solution structure of the 2nd PHD domain from Mi2b | | Descriptor: | Mi2-beta, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
1MM3
 
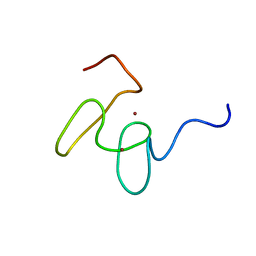 | | Solution structure of the 2nd PHD domain from Mi2b with C-terminal loop replaced by corresponding loop from WSTF | | Descriptor: | Mi2-beta(Chromodomain helicase-DNA-binding protein 4) and transcription factor WSTF, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
1N0Z
 
 | | Solution structure of the first zinc-finger domain from ZNF265 | | Descriptor: | ZINC ION, ZNF265 | | Authors: | Plambeck, C.A, Fairley, K, Kwan, A.H.Y, Westman, B.J, Adams, D, Morris, B, Mackay, J.P. | | Deposit date: | 2002-10-15 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the zinc finger domain from human splicing factor ZNF265 fold
J.BIOL.CHEM., 278, 2003
|
|
1Y0J
 
 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1M36
 
 | |
1LIQ
 
 | | Non-native Solution Structure of a fragment of the CH1 domain of CBP | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Matthews, J.M, Kwan, A.H.Y, Newton, A, Gell, D.A, Crossley, M, Mackay, J.P. | | Deposit date: | 2002-04-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Zinc Binding Fold Underlines the Versatility of Zinc Binding Modules in Protein Evolution
Structure, 10, 2002
|
|
1Y01
 
 | | Crystal structure of AHSP bound to Fe(II) alpha-hemoglobin | | Descriptor: | 6-[(CYCLOHEXYLACETYL)(2-HYDROXYETHYL)AMINO]-6-DEOXY-D-XYLO-HEXITOL, Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, ... | | Authors: | Feng, L, Gell, D.A, Zhou, S, Gu, L, Gow, A.J, Weiss, M.J, Mackay, J.P, Shi, Y. | | Deposit date: | 2004-11-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin.
Cell(Cambridge,Mass.), 119, 2004
|
|
1Y04
 
 | | Solution structure of a recombinant type I sculpin antifreeze protein | | Descriptor: | Antifreeze peptide SS-3 | | Authors: | Kwan, A.H.Y, Fairley, K, Anderberg, P.I, Liew, C.W, Harding, M.M, Mackay, J.P. | | Deposit date: | 2004-11-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant type I sculpin antifreeze protein
Biochemistry, 44, 2005
|
|
1Z8U
 
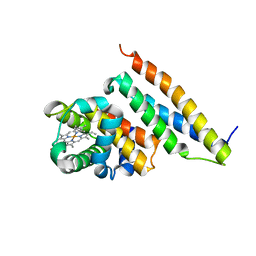 | | Crystal structure of oxidized alpha hemoglobin bound to AHSP | | Descriptor: | Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Feng, L, Zhou, S, Gu, L, Gell, D.A, Mackay, J.P, Weiss, M.J, Gow, A.J, Shi, Y. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem.
Nature, 435, 2005
|
|
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | Descriptor: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | Authors: | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
1U86
 
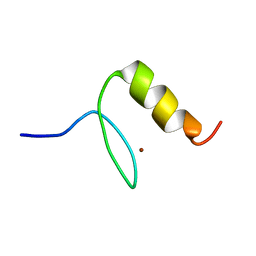 | |
1WO3
 
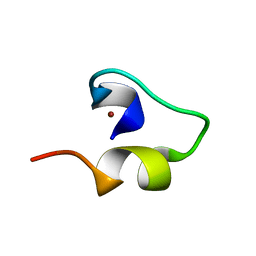 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO6
 
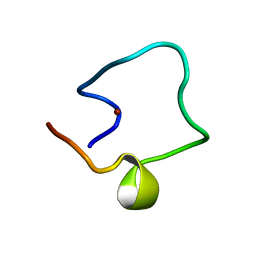 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO4
 
 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
