4FCM
 
 | |
4FCJ
 
 | |
3C9N
 
 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
3UJM
 
 | |
1XWV
 
 | | Structure of the house dust mite allergen Der f 2: Implications for function and molecular basis of IgE cross-reactivity | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Der f II | | Authors: | Johannessen, B.R, Skov, L.K, Kastrup, J.S, Kristensen, O, Bolwig, C, Larsen, J.N, Spangfort, M, Lund, K, Gajhede, M. | | Deposit date: | 2004-11-02 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the house dust mite allergen Der f 2: implications for function and molecular basis of IgE cross-reactivity.
Febs Lett., 579, 2005
|
|
1XR9
 
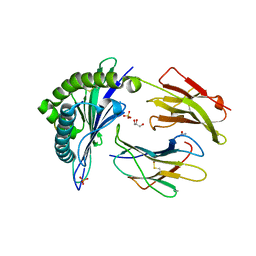 | | Crystal Structures of HLA-B*1501 in Complex with Peptides from Human UbcH6 and Epstein-Barr Virus EBNA-3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G, Blicher, T, Johannessen, B.R, Kristensen, O, Buus, S, Gajhede, M. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1XR8
 
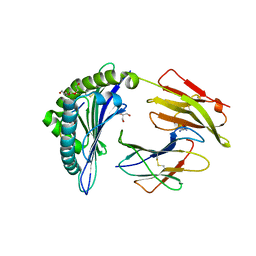 | | Crystal Structures of HLA-B*1501 in Complex with Peptides from Human UbcH6 and Epstein-Barr Virus EBNA-3 | | Descriptor: | Beta-2-microglobulin, EBNA-3 nuclear protein, GLYCEROL, ... | | Authors: | Roder, G, Blicher, T, Johannessen, B.R, Kristensen, O, Buus, S, Gajhede, M. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1FEU
 
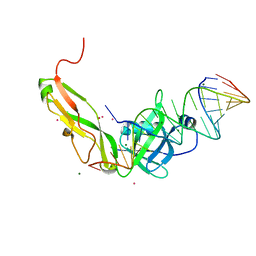 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FNM
 
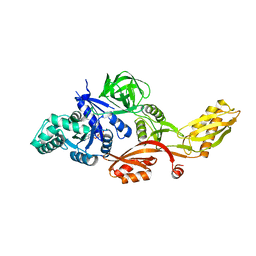 | | STRUCTURE OF THERMUS THERMOPHILUS EF-G H573A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Laurberg, M, Kristensen, O, Martemyanov, K, Gudkov, A.T, Nagaev, I, Hughes, D, Liljas, A. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site.
J.Mol.Biol., 303, 2000
|
|
1LOU
 
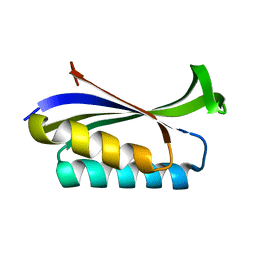 | | RIBOSOMAL PROTEIN S6 | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E, Kristensen, O, Proctor, M, Oliveberg, M. | | Deposit date: | 1998-11-25 | | Release date: | 1998-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural changes in the transition state of protein folding: alternative interpretations of curved chevron plots.
Biochemistry, 38, 1999
|
|
4CBP
 
 | | Crystal structure of neural ectodermal development factor IMP-L2. | | Descriptor: | GLYCEROL, NEURAL/ECTODERMAL DEVELOPMENT FACTOR IMP-L2 | | Authors: | Kulahin, N, Kristensen, O, Brzozowski, M, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis of Imp-L2 Function
To be Published
|
|
2WL7
 
 | | Crystal structure of the PSD93 PDZ1 domain | | Descriptor: | CHLORIDE ION, DISKS LARGE HOMOLOG 2, SULFATE ION | | Authors: | Fiorentini, M, Kallehauge, A, Kristensen, O, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2009-06-22 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of the First Pdz Domain of Human Psd-93.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2XYC
 
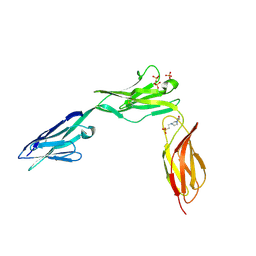 | | CRYSTAL STRUCTURE OF NCAM2 IGIV-FN3I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NEURAL CELL ADHESION MOLECULE 2, ... | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2WIM
 
 | | Crystal structure of NCAM2 IG1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Kristensen, O, Rasmussen, K, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural model and trans-interaction of the entire ectodomain of the olfactory cell adhesion molecule.
Structure, 19, 2011
|
|
2XY1
 
 | | CRYSTAL STRUCTURE OF NCAM2 IG3-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2XY2
 
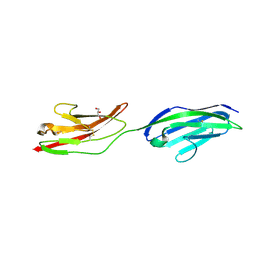 | | CRYSTAL STRUCTURE OF NCAM2 IG1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2J3P
 
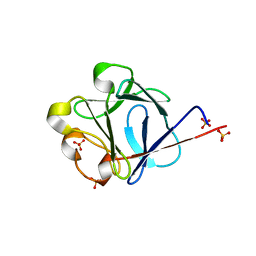 | | crystal structure of rat FGF1 at 1.4 A | | Descriptor: | HEPARIN-BINDING GROWTH FACTOR 1, SULFATE ION | | Authors: | Kulahin, N, Kristensen, O, Berezin, V, Gajhede, M, Bock, E. | | Deposit date: | 2006-08-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Rat Acidic Fibroblast Growth Factor at 1.4 A Resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2J06
 
 | |
2J05
 
 | |
2JLL
 
 | | Crystal structure of NCAM2 IgIV-FN3II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Kulahin, N, Rasmussen, K, Kristensen, O, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | Deposit date: | 2008-09-10 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
3FYS
 
 | | Crystal Structure of DegV, a fatty acid binding protein from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, PALMITIC ACID, ... | | Authors: | Nan, J, Zhou, Y.F, Yang, C. | | Deposit date: | 2009-01-23 | | Release date: | 2009-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a fatty acid-binding protein from Bacillus subtilis determined by sulfur-SAD phasing using in-house chromium radiation
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1XKG
 
 | | Crystal structure of the major house dust mite allergen Der p 1 in its pro form at 1.61 A resolution | | Descriptor: | GLYCEROL, Major mite fecal allergen Der p 1, SULFATE ION, ... | | Authors: | Meno, K, Thorsted, P.B, Gajhede, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The crystal structure of recombinant proDer p 1, a major house dust mite proteolytic allergen.
J.Immunol., 175, 2005
|
|
1JQS
 
 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQM
 
 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
2DTM
 
 | | Thermodynamic and structural analyses of hydrolytic mechanism by catalytic antibodies | | Descriptor: | IMMUNOGLOBULIN 6D9 | | Authors: | Oda, M, Ito, N, Tsumuraya, T, Suzuki, K, Fujii, I. | | Deposit date: | 2006-07-13 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
