5GJM
 
 | |
5GJP
 
 | |
7DIB
 
 | |
6JTT
 
 | | MHETase in complex with BHET | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, ... | | Authors: | Sagong, H.-Y, Seo, H, Kim, K.-J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
5HXX
 
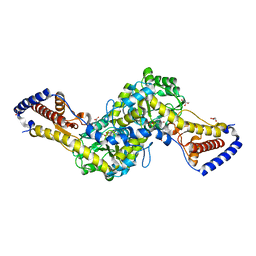 | | Crystal structure of AspAT from Corynebacterium glutamicum | | Descriptor: | 2-OXOGLUTARIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, ... | | Authors: | Son, H.-F, Kim, K.-J. | | Deposit date: | 2016-01-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into a novel class of aspartate aminotransferase from Corynebacterium glutamicum
To be published
|
|
6JTU
 
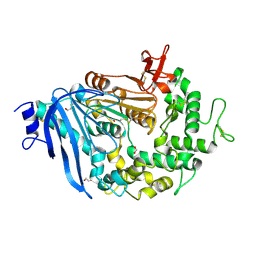 | | Crystal structure of MHETase from Ideonella sakaiensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Sagong, H.-Y, Seo, H, Kim, K.-J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
5GJO
 
 | |
5IJZ
 
 | |
8I4R
 
 | |
7CBE
 
 | |
7FBG
 
 | |
6IJK
 
 | |
6J2U
 
 | |
6KUS
 
 | |
6KUQ
 
 | |
6L3O
 
 | |
6KUO
 
 | |
6AG8
 
 | |
5YNS
 
 | |
5X5H
 
 | | Crystal structure of metB from Corynebacterium glutamicum | | Descriptor: | Cystathionine beta-lyases/cystathionine gamma-synthases, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insights into Substrate Specificity of Cystathionine gamma-Synthase from Corynebacterium glutamicum
J. Agric. Food Chem., 65, 2017
|
|
5XJH
 
 | |
7C11
 
 | | Formate--tetrahydrofolate ligase from Methylobacterium extorquens CM4 strain | | Descriptor: | ACETATE ION, CITRATE ANION, Formate-tetrahydrofolate ligase, ... | | Authors: | Kim, K.-J, Kim, S, Seo, H, Lee, S. | | Deposit date: | 2020-05-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.815 Å) | | Cite: | Biochemical properties and crystal structure of formate-tetrahydrofolate ligase from Methylobacterium extorquens CM4.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
8JCT
 
 | |
5M47
 
 | |
4REP
 
 | | Crystal Structure of gamma-carotenoid desaturase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Gamma-carotene desaturase | | Authors: | Ahn, J.-W, Kim, E.-J, Kim, S, Kim, K.-J. | | Deposit date: | 2014-09-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of 1'-OH-carotenoid 3,4-desaturase from Nonlabens dokdonensis DSW-6.
Enzyme.Microb.Technol., 77, 2015
|
|
