8HIQ
 
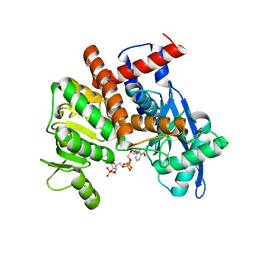 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
8HIZ
 
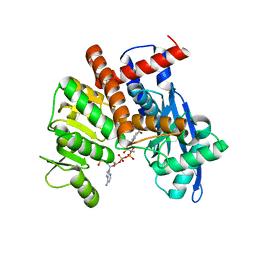 | | cryoEM structure of glutamate dehydrogenase from Thermococcus profundus in complex with NADP | | Descriptor: | Glutamate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wakabayashi, T, Oide, M, Kato, T, Nakasako, M. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Coenzyme-binding pathway on glutamate dehydrogenase suggested from multiple-binding sites visualized by cryo-electron microscopy.
Febs J., 290, 2023
|
|
3WO9
 
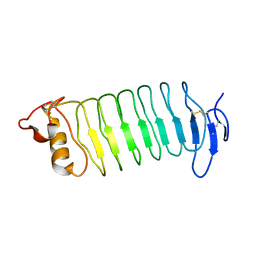 | | Crystal structure of the lamprey variable lymphocyte receptor C | | Descriptor: | Variable lymphocyte receptor C | | Authors: | Kanda, R, Sutoh, Y, Kasamatsu, J, Maenaka, K, Kasahara, M, Ose, T. | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the lamprey variable lymphocyte receptor C reveals an unusual feature in its N-terminal capping module.
Plos One, 9, 2014
|
|
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5X02
 
 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor FF-10101 | | Descriptor: | N-[(2S)-1-[5-[2-[(4-cyanophenyl)amino]-4-(propylamino)pyrimidin-5-yl]pent-4-ynylamino]-1-oxidanylidene-propan-2-yl]-4-(dimethylamino)-N-methyl-but-2-enamide, Receptor-type tyrosine-protein kinase FLT3, SULFATE ION | | Authors: | Fujikawa, N, Hirano, D, Takasaki, M, Terada, D, Hagiwara, S, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells withFLT3mutations.
Blood, 131, 2018
|
|
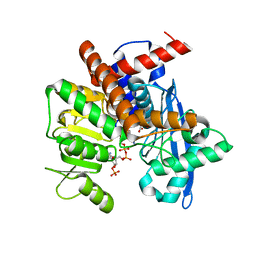
8XCR
 
 | |
8XCV
 
 | |
8XCW
 
 | |
8XD0
 
 | |
8XD1
 
 | |
8XD5
 
 | |
8XCQ
 
 | |
8XCU
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus incorporating NADPH, AKG and GLU in the steady stage of reaction | | Descriptor: | 2-OXOGLUTARIC ACID, GAMMA-L-GLUTAMIC ACID, Glutamate dehydrogenase, ... | | Authors: | Wakabayashi, T, Oide, M, Nakasako, M. | | Deposit date: | 2023-12-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
8XCX
 
 | |
8XD4
 
 | |
8XD2
 
 | |
8XCO
 
 | |
8XCS
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus in complex with NADPH, AKG and NH4 in the initial stage of reaction | | Descriptor: | 2-OXOGLUTARIC ACID, AMMONIUM ION, Glutamate dehydrogenase, ... | | Authors: | Wakabayashi, T, Oide, M, Nakasako, M. | | Deposit date: | 2023-12-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
8XCY
 
 | |
8XD6
 
 | | Cryo-EM structure of Glutamate dehydrogenase from Thermococcus profundus in complex with NADPH, AKG and NH4 in the steady stage of reaction | | Descriptor: | 2-OXOGLUTARIC ACID, AMMONIUM ION, Glutamate dehydrogenase, ... | | Authors: | Wakabayashi, T, Oide, M, Nakasako, M. | | Deposit date: | 2023-12-10 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM-sampling of metastable conformations appearing in cofactor-ligand association and catalysis of glutamate dehydrogenase.
Sci Rep, 14, 2024
|
|
8XD3
 
 | |
8XCP
 
 | |
8XCT
 
 | |
8XCZ
 
 | |
5LQF
 
 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
