5JOD
 
 | | Structure of proplasmepsin IV from Plasmodium falciparum | | Descriptor: | GLYCEROL, Proplasmepsin IV | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2KZF
 
 | | Solution NMR structure of the thermotoga maritima protein TM0855 a putative ribosome binding factor A | | Descriptor: | Ribosome-binding factor A | | Authors: | Serrano, P, Jaudzems, K, Horst, R, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the thermotoga maritima protein TM0855
To be Published
|
|
2KYS
 
 | | NMR Structure of the SARS Coronavirus Nonstructural Protein Nsp7 in Solution at pH 6.5 | | Descriptor: | Non-structural protein 7 | | Authors: | Johnson, M.A, Jaudzems, K, Wilson, I.A, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the SARS-CoV Nonstructural Protein 7 in Solution at pH 6.5.
J.Mol.Biol., 402, 2010
|
|
2KTS
 
 | | NMR structure of the protein NP_415897.1 | | Descriptor: | Heat shock protein hslJ | | Authors: | Serrano, P, Jaudzems, K, Geralt, M, Horst, R, Wuthrich, K, Wilson, I.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-02-06 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_415897.1
To be Published
|
|
2LTH
 
 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | Deposit date: | 2012-05-25 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
2MX9
 
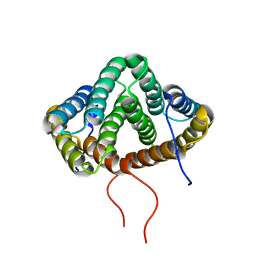 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 5.5 | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-19 | | Method: | SOLUTION NMR | | Cite: | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2MX8
 
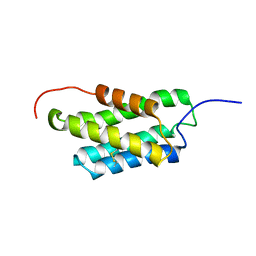 | | NMR structure of N-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) at pH 7.2 | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Chen, G, Nordling, K, Rising, A, Johansson, J. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Diversified Structural Basis of a Conserved Molecular Mechanism for pH-Dependent Dimerization in Spider Silk N-Terminal Domains.
Chembiochem, 16, 2015
|
|
2MFZ
 
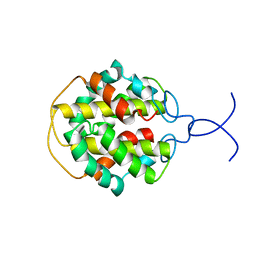 | | NMR structure of C-terminal domain from A. ventricosus minor ampullate spidroin (MiSp) | | Descriptor: | Minor ampullate spidroin | | Authors: | Otikovs, M, Jaudzems, K, Andersson, M, Chen, G, Landreh, M, Nordling, K, Kronqvist, N, Westermark, P, Jornvall, H, Knight, S, Ridderstrale, Y, Holm, L, Meng, Q, Chesler, M, Johansson, J, Rising, A. | | Deposit date: | 2013-10-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbonic Anhydrase Generates CO2 and H+ That Drive Spider Silk Formation Via Opposite Effects on the Terminal Domains
Plos Biol., 12, 2014
|
|
2K7R
 
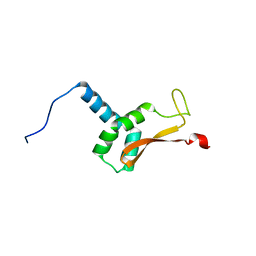 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
4FBS
 
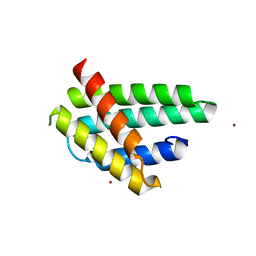 | | Structure of monomeric NT from Euprosthenops australis Major Ampullate Spidroin 1 (MaSp1) | | Descriptor: | BROMIDE ION, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, M, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain
J.Mol.Biol., 2012
|
|
2QTD
 
 | |
4D53
 
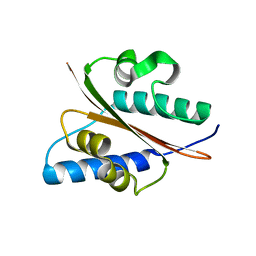 | | Outer surface protein BB0689 from Borrelia burgdorferi | | Descriptor: | BB0689 | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2014-11-02 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Analysis of Bb0689 from Borrelia Burgdorferi, a Member of the Bacterial CAP Superfamily.
J.Struct.Biol., 192, 2015
|
|
6R1G
 
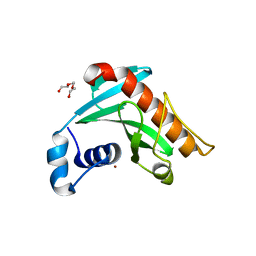 | | Crystal structure of Borrelia burgdorferi periplasmic protein BB0365 (IPLA7, p22) | | Descriptor: | Outer surface 22 kDa lipoprotein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Borrelia burgdorferi periplasmic lipoprotein BB0365 involved in Lyme disease infection.
Febs Lett., 594, 2020
|
|
5FS4
 
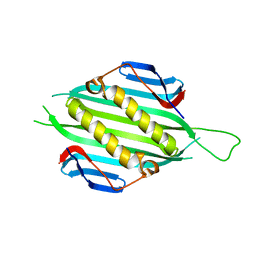 | | Bacteriophage AP205 coat protein | | Descriptor: | AP205 BACTERIOPHAGE COAT PROTEIN | | Authors: | Shishovs, M, Tars, K. | | Deposit date: | 2015-12-29 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of Ap205 Coat Protein Reveals Circular Permutation in Ssrna Bacteriophages.
J.Mol.Biol., 428, 2016
|
|
5NDA
 
 | | NMR Structural Characterisation of Pharmaceutically Relevant Proteins Obtained Through a Novel Recombinant Production: The Case of The Pulmonary Surfactant Polypeptide C Analogue rSP-C33Leu. | | Descriptor: | rSP-C33Leu -RECOMBINANT PULMONARY SURFACTANT-ASSOCIATED POLYPEPTIDE C ANALOGUE- | | Authors: | Venturi, L, Pioselli, B, Johansson, J, Rising, A, Kronqvist, N, Nordling, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Efficient protein production inspired by how spiders make silk.
Nat Commun, 8, 2017
|
|
8OG0
 
 | |
6FXE
 
 | | BBE31 from Lyme disease agent Borrelia (Borreliella) burgdorferi playing a vital role in successful colonization of the mammalian host | | Descriptor: | GLUTATHIONE, Putative surface protein | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
6FZE
 
 | | BBE31 from Lyme disease agent Borrelia (Borreliella) burgdorferi playing a vital role in successful colonization of the mammalian host (native data) | | Descriptor: | GLUTATHIONE, Putative surface protein, SULFATE ION | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
6FZZ
 
 | | Crystal structure of BSE31 (BSPA14S_RS05060 gene product) from Lyme disease agent Borrelia (Borreliella) spielmanii | | Descriptor: | TETRAETHYLENE GLYCOL, Virulent strain associated lipoprotein | | Authors: | Brangulis, K, Akopjana, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BBE31 from the Lyme disease agent Borrelia burgdorferi, known to play an important role in successful colonization of the mammalian host, shows the ability to bind glutathione.
Biochim Biophys Acta Gen Subj, 1864, 2019
|
|
4BG0
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Baumanis, V, Tars, K. | | Deposit date: | 2013-03-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
4CBE
 
 | | Crystal structure of complement factors H and FHL-1 binding protein BBH06 or CRASP-2 from Borrelia burgdorferi (Native) | | Descriptor: | COMPLEMENT REGULATOR-ACQUIRING SURFACE PROTEIN 2 (CRASP-2) | | Authors: | Brangulis, K, Petrovskis, I, Kazaks, A, Ranka, R, Tars, K. | | Deposit date: | 2013-10-13 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterization of Cspz, a Complement Regulator Factor H and Fhl-1 Binding Protein from Borrelia Burgdorferi.
FEBS J., 281, 2014
|
|
8PJ0
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-3-methylbutanamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-06-22 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
5LQP
 
 | |
8RZE
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
