1VTR
 
 | | STRUCTURE OF THE DEOXYTETRANUCLEOTIDE D-PAPTPAPT AND A SEQUENCE-DEPENDENT MODEL FOR POLY(DA-DT) | | Descriptor: | DNA (5'-D(*AP*TP*AP*T)-3') | | Authors: | Viswamitra, M.A, Shakked, Z, Jones, P.G, Sheldrick, G.M, Salisbury, S.A, Kennard, O. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of the Deoxytetranucleotide d-pApTpApT and a Sequence-Dependent Model for Poly(dA-dT)
Biopolymers, 21, 1982
|
|
1RNB
 
 | |
1BGS
 
 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
1BAN
 
 | |
1B27
 
 | |
1B2U
 
 | |
1B2S
 
 | |
1B3S
 
 | |
1BAO
 
 | |
1BRS
 
 | |
1BSE
 
 | |
1BSA
 
 | |
1BNJ
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 9.0 | | Descriptor: | BARNASE | | Authors: | Cameron, A, Henrick, K, Fersht, A.R, Dodson, G, Buckle, A.M. | | Deposit date: | 1995-05-17 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structural analysis of mutations in the hydrophobic cores of barnase.
J.Mol.Biol., 234, 1993
|
|
1BSD
 
 | |
1B21
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B2X
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 7.5 FROM A CRYO_COOLED CRYSTAL AT 100K | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Harrison, P, Vaughan, C.K, Buckle, A.M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B20
 
 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B2Z
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BNS
 
 | | STRUCTURAL STUDIES OF BARNASE MUTANTS | | Descriptor: | BARNASE | | Authors: | Chen, Y.W. | | Deposit date: | 1994-04-11 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of buried hydrogen bonds to protein stability. The crystal structures of two barnase mutants.
J.Mol.Biol., 234, 1993
|
|
7O70
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4R,7S)-12-chloro-14-fluoro-13-(2-fluoro-6-hydroxyphenyl)-4-methyl-10-oxa-2,5,16,18-tetrazatetracyclo[9.7.1.0^(2,7).0^(15,19)]nonadeca-1(18),11,13,15(19),16-pentaen-5-en-1-one-yl]prop-2, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7O83
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(7S)-11-chloro-12-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,15,17-tetrazatetracyclo[8.7.1.02,7.014,18]octadeca-1(17),10,12,14(18),15-pentaen-5-yl]prop-2-en-1-one, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7OLX
 
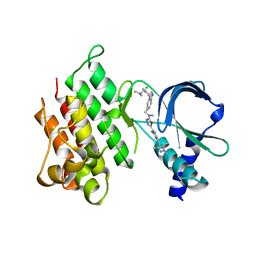 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLS
 
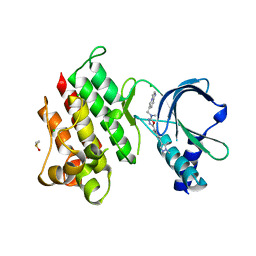 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLV
 
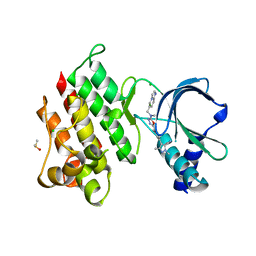 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl, cyano pyrazole group | | Descriptor: | 4-[4-[5-[imidazo[1,2-a]pyridin-6-ylmethyl(methyl)amino]-1,3,4-oxadiazol-2-yl]-3-methyl-phenyl]-2,5-dimethyl-pyrazole-3-carbonitrile, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OO7
 
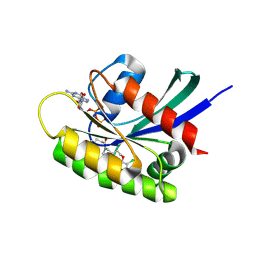 | | KRasG12C ligand complex | | Descriptor: | 1-[(6aS)-3-chloro-2-(5-methyl-1H-indazol-4-yl)-5,6,6a,7,9,10-hexahydro-8H-pyrazino[1',2':5,6][1,5]oxazocino[4,3,2-de]quinazolin-8-yl]-2-propen-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-05-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
