7PCM
 
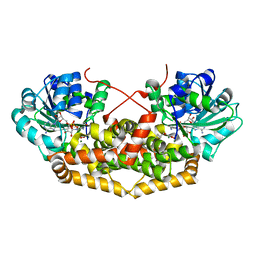 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCN
 
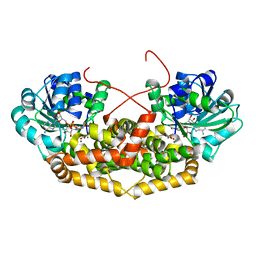 | | BurG (holo) in complex with gonyenediol (14), trigonic acid (6) and DMS: Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R)-2-oxidanyl-2-(1-oxidanylcyclopropyl)ethanoic acid, (METHYLSULFANYL)METHANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCT
 
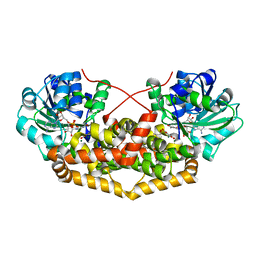 | | BurG E232Q mutant (holo) in complex with enol-oxalacetate (15): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
5MR6
 
 | | XiaF from Streptomyces sp. in complex with FADH2 and Glycerol | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, XiaF protein | | Authors: | Kugel, S, Baunach, M, Baer, P, Ishida-Ito, M, Sundaram, S, Xu, Z, Groll, M, Hertweck, C. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptic indole hydroxylation by a non-canonical terpenoid cyclase parallels bacterial xenobiotic detoxification.
Nat Commun, 8, 2017
|
|
5C1J
 
 | |
7AYY
 
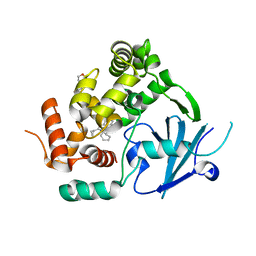 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
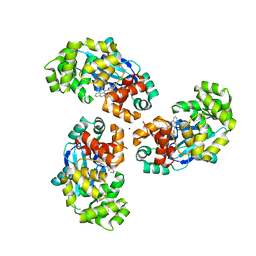 | |
7AZ0
 
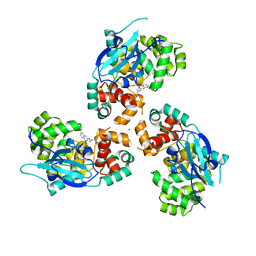 | |
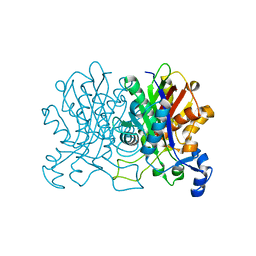
4YUF
 
 | |
4YUC
 
 | | Crystal Structure of CorB derivatized with S-(2-acetamidoethyl) 4-methyl-3-oxohexanethioate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CorB, SODIUM ION | | Authors: | Zocher, G, Vilstrup, J, Stehle, T. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis of head to head polyketide fusion by CorB.
Chem Sci, 6, 2015
|
|
4KC5
 
 | |
5LX7
 
 | | Cys-Gly dipeptidase GliJ mutant D38N | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dipeptidase, FE (III) ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX0
 
 | | Cys-Gly dipeptidase GliJ (space group P3221) | | Descriptor: | Dipeptidase, FE (III) ION | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX1
 
 | | Cys-Gly dipeptidase GliJ mutant D304A | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LX4
 
 | | Cys-Gly dipeptidase GliJ mutant D38H | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5LWZ
 
 | | Cys-Gly dipeptidase GliJ (space group C2) | | Descriptor: | Dipeptidase, FE (III) ION, GLYCEROL | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRU
 
 | | Cys-Gly dipeptidase GliJ in complex with Zn2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, ZINC ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRX
 
 | | Cys-Gly dipeptidase GliJ in complex with Fe2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS2
 
 | | Cys-Gly dipeptidase GliJ in complex with Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRT
 
 | | Cys-Gly dipeptidase GliJ in complex with Ca2+ | | Descriptor: | CALCIUM ION, Dipeptidase gliJ, MAGNESIUM ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS1
 
 | | Cys-Gly dipeptidase GliJ in complex with Ni2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, NICKEL (II) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRZ
 
 | | Cys-Gly dipeptidase GliJ in complex with Mn2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, GLYCEROL, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS5
 
 | | Cys-Gly dipeptidase GliJ in complex with Cu2+ and Zn2+ | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRY
 
 | | Cys-Gly dipeptidase GliJ in complex with Fe3+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
7NCL
 
 | | Glutathione-S-transferase GliG mutant E82Q | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glutathione S-transferase GliG | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insights into C-S Bond Formation in Gliotoxin.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
