7KLK
 
 | | Human Arginase1 Complexed with Inhibitor Compound 3a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-carboxypiperidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
3L5B
 
 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L59
 
 | | Structure of BACE Bound to SCH710413 | | Descriptor: | (2Z)-3-(3-chlorobenzyl)-2-imino-5,5-dimethylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L58
 
 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5E
 
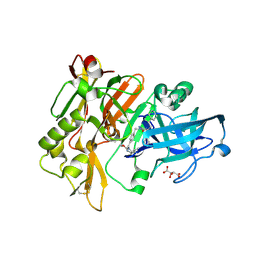 | | Structure of BACE Bound to SCH736062 | | Descriptor: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5D
 
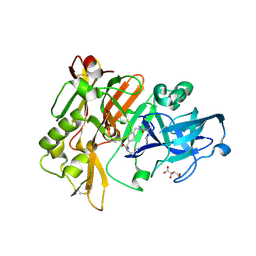 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
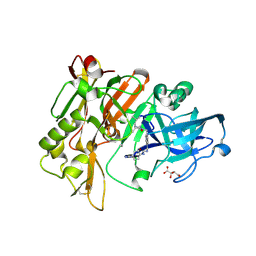 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5F
 
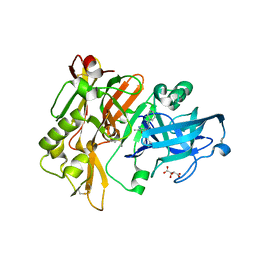 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
5C45
 
 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6M)-2-[(3S)-1-{[2-(methylamino)pyrimidin-5-yl]methyl}piperidin-3-yl]-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Selective small-molecule inhibition of an RNA structural element.
Nature, 526, 2015
|
|
3GIP
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | Descriptor: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3GIQ
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc and phosphonate inhibitor, a mimic of the reaction tetrahedral intermediate. | | Descriptor: | N-[(R)-hydroxy(methyl)phosphoryl]-D-glutamic acid, N-acyl-D-glutamate deacylase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3ID7
 
 | | Crystal structure of renal dipeptidase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-07-20 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
8CIJ
 
 | | CRYSTAL STRUCTURE OF HUMAN HPK1 (MAP4K1) COMPLEX WITH 2-[8-Amino-7-fluoro-6-(8-methyl-2,3-dihydro-1H-pyrido[2,3-b][1,4]oxazin-7-yl)-isoquinolin-3-ylamino]-6-isopropyl-5,6-dihydro-4H-1,6,8a-triaza-azulen-7-one | | Descriptor: | 2-[[8-azanyl-7-fluoranyl-6-(8-methyl-2,3-dihydro-1~{H}-pyrido[2,3-b][1,4]oxazin-7-yl)isoquinolin-3-yl]amino]-6-propan-2-yl-5,8-dihydro-4~{H}-pyrazolo[1,5-d][1,4]diazepin-7-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Musil, D, Toure, M. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Discovery of quinazoline HPK1 inhibitors with high cellular potency.
Bioorg.Med.Chem., 92, 2023
|
|
8CDW
 
 | |
3K5X
 
 | | Crystal structure of dipeptidase from Streptomics coelicolor complexed with phosphinate pseudodipeptide L-Ala-D-Asp at 1.4A resolution. | | Descriptor: | Dipeptidase, ZINC ION, phosphinate pseudodipeptide L-Ala-D-Asp | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
Biochemistry, 49, 2010
|
|
1AHJ
 
 | | NITRILE HYDRATASE | | Descriptor: | FE (III) ION, NITRILE HYDRATASE (SUBUNIT ALPHA), NITRILE HYDRATASE (SUBUNIT BETA) | | Authors: | Huang, W, Schneider, G, Lindqvist, Y. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold.
Structure, 5, 1997
|
|
5KX9
 
 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6P)-2-{(3S)-1-[(2-methoxypyrimidin-5-yl)methyl]piperidin-3-yl}-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atomic resolution mechanistic studies of ribocil: A highly selective unnatural ligand mimic of the E. coli FMN riboswitch.
Rna Biol., 13, 2016
|
|
