6JCR
 
 | | AAV1 in neutral condition at 3.07 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCQ
 
 | | AAV1 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCT
 
 | | AAV5 in neutral condition at 3.18 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6IHB
 
 | | Adeno-Associated Virus 2 in complex with AAVR | | Descriptor: | Capsid protein VP1, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
8XAI
 
 | |
6IH9
 
 | | Adeno-Associated Virus 2 at 2.8 ang | | Descriptor: | Capsid protein VP1 | | Authors: | Lou, Z.Y, Zhang, R. | | Deposit date: | 2018-09-29 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Adeno-associated virus 2 bound to its cellular receptor AAVR.
Nat Microbiol, 4, 2019
|
|
8VTT
 
 | |
8VTS
 
 | |
7KGE
 
 | |
7KGI
 
 | |
7KGF
 
 | |
7KGH
 
 | |
7KGD
 
 | |
7KGG
 
 | |
6ALE
 
 | | A V-to-F substitution in SK2 channels causes Ca2+ hypersensitivity and improves locomotion in a C. elegans ALS model | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin-2, ... | | Authors: | Nam, Y.W, Zhang, M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A V-to-F substitution in SK2 channels causes Ca2+hypersensitivity and improves locomotion in a C. elegans ALS model.
Sci Rep, 8, 2018
|
|
8TZQ
 
 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE Mutant, scFv16, and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-27 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
8U02
 
 | | CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine | | Descriptor: | D(2) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Krumm, B.E, Kapolka, N.J, Fay, J.F, Roth, B.L. | | Deposit date: | 2023-08-28 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A neurodevelopmental disorder mutation locks G proteins in the transitory pre-activated state.
Nat Commun, 15, 2024
|
|
7XDQ
 
 | | Crystal structure of a glucosylglycerol phosphorylase mutant from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase, LITHIUM ION, beta-D-glucopyranose | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7XDR
 
 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
1H9C
 
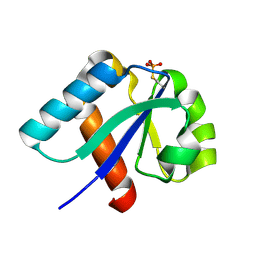 | | NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N'-diacetylchitobiose specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. | | Descriptor: | PTS SYSTEM, CHITOBIOSE-SPECIFIC IIB COMPONENT | | Authors: | Ab, E, Schuurman-Wolters, G.K, Nijlant, D, Dijkstra, K, Saier, M.H, Robillard, G.T, Scheek, R.M. | | Deposit date: | 2001-03-07 | | Release date: | 2001-05-21 | | Last modified: | 2018-01-31 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Cysteinyl-Phosphorylated Enzyme Iib of the N,N'-Diacetylchitobiose Specific Phosphoenolpyruvate-Dependentphosphotransferase System of Escherichia Coli
J.Mol.Biol., 308, 2001
|
|
1E2B
 
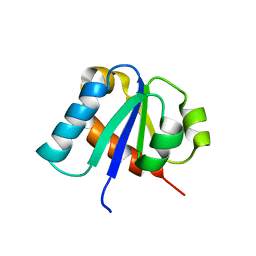 | | NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES | | Descriptor: | ENZYME IIB-CELLOBIOSE | | Authors: | Ab, E, Schuurman-Wolters, G, Reizer, J, Saier, M.H, Dijkstra, K, Scheek, R.M, Robillard, G.T. | | Deposit date: | 1996-11-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR side-chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli.
Protein Sci., 6, 1997
|
|
