5TGN
 
 | | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus
To Be Published
|
|
5TJJ
 
 | | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, Transcriptional regulator, IclR family | | Authors: | Michalska, K, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius
To Be Published
|
|
1YZF
 
 | | Crystal structure of the lipase/acylhydrolase from Enterococcus faecalis | | Descriptor: | lipase/acylhydrolase | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lipase/acylhydrolase from Enterococcus faecalis
To be Published
|
|
1Z0P
 
 | | Crystal structure of the Protein of Unknown Function SPY1572 from Streptococcus pyogenes | | Descriptor: | hypothetical protein SPy1572 | | Authors: | Zhang, R, Lezondra, L, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-02 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7A Crystal structure of the hypothetical protein SPy1572 from Streptococcus pyogenes
To be Published
|
|
1Z67
 
 | | Structure of Homeodomain-like Protein of Unknown Function S4005 from Shigella flexneri | | Descriptor: | SODIUM ION, hypothetical protein S4005 | | Authors: | Osipiuk, J, Maltseva, N, Dementieva, I, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of YidB protein from Shigella flexneri shows a new fold with homeodomain motif.
Proteins, 65, 2006
|
|
2NPN
 
 | | Crystal structure of putative cobalamin synthesis related protein (CobF) from Corynebacterium diphtheriae | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative cobalamin synthesis related protein, ... | | Authors: | Nocek, B, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of cobalamin synthesis related protein (CobF) from Corynebacterium diphtheriae
To be Published
|
|
2OCZ
 
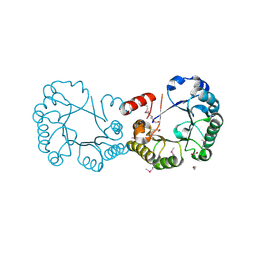 | | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes. | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes.
TO BE PUBLISHED
|
|
4XLT
 
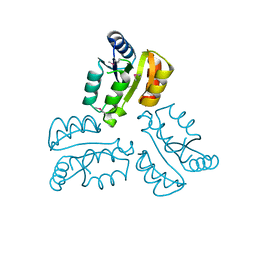 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | Response regulator receiver protein | | Authors: | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
4XS5
 
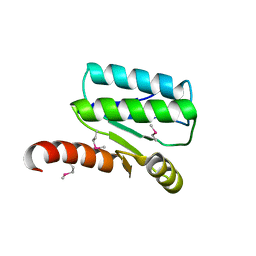 | | Crystal structure of Sulfate transporter/antisigma-factor antagonist STAS from Dyadobacter fermentans DSM 18053 | | Descriptor: | Sulfate transporter/antisigma-factor antagonist STAS | | Authors: | Chang, C, Cuff, M, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Sulfate transporter/antisigma-factor antagonist STAS from Dyadobacter fermentans DSM 18053
To Be Published
|
|
4ZPJ
 
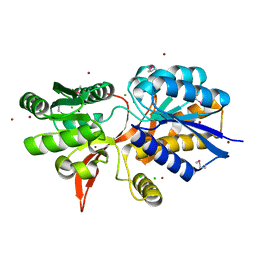 | | ABC transporter substrate-binding protein from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, Extracellular ligand-binding receptor, ZINC ION | | Authors: | OSIPIUK, J, Holowicki, J, Clancy, S, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | ABC transporter substrate-binding protein from Sphaerobacter thermophilus.
to be published
|
|
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | Descriptor: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
4YCS
 
 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
6V72
 
 | | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase II, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis
To Be Published
|
|
6V70
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Cadmium in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CADMIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6V54
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6V61
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
To Be Published
|
|
6V5M
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in Complex with Succinate.
To Be Published
|
|
6V71
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
6V73
 
 | | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis with beta mercaptoethanol in the active site | | Descriptor: | BETA-MERCAPTOETHANOL, Beta-lactamase II, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Erythrobacter litoralis with beta mercaptoethanol in the active site
To Be Published
|
|
3TT2
 
 | | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GCN5-related N-acetyltransferase, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of GCN5-related N-Acetyltransferase from Sphaerobacter thermophilus
To be Published
|
|
3TOV
 
 | |
3SHP
 
 | | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | Putative acetyltransferase Sthe_0691, S,R MESO-TARTARIC ACID | | Authors: | Chang, C, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745
To be Published
|
|
3V7B
 
 | | Dip2269 protein from corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein | | Authors: | Osipiuk, J, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Dip2269 protein from corynebacterium diphtheriae.
To be Published
|
|
3SJR
 
 | |
