4JJB
 
 | |
4J9H
 
 | |
4J9I
 
 | |
4J9F
 
 | |
4J9G
 
 | |
4JJD
 
 | |
4J9C
 
 | |
4JJC
 
 | |
4F16
 
 | |
4F17
 
 | |
3V58
 
 | |
1KBY
 
 | | Structure of Photosynthetic Reaction Center with bacteriochlorophyll-bacteriopheophytin heterodimer | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C, Goetsch, A, Allen, J.P. | | Deposit date: | 2001-11-07 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of the heterodimer reaction center from Rhodobacter sphaeroides at 2.55 a resolution.
Photosynth.Res., 74, 2002
|
|
3V57
 
 | |
5I11
 
 | |
5IHK
 
 | |
5IHN
 
 | |
5IHI
 
 | |
2HKI
 
 | |
5OB1
 
 | |
5OAV
 
 | |
5OAZ
 
 | |
5OB0
 
 | |
5OB2
 
 | |
1JDL
 
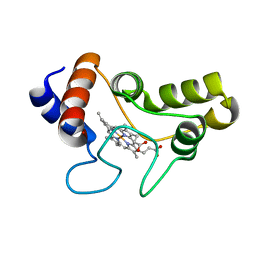 | | Structure of cytochrome c2 from Rhodospirillum Centenum | | Descriptor: | CYTOCHROME C2, ISO-2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Camara-Artigas, A, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-14 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of cytochrome c2 from Rhodospirillum centenum.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JGZ
 
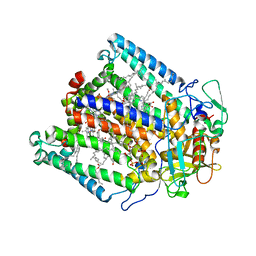 | | Photosynthetic Reaction Center Mutant With Tyr M 76 Replaced With Lys | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Camara-Artigas, A, Magee, C.L, Williams, J.C, Allen, J.P. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Individual interactions influence the crystalline order for membrane proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
