6WMR
 
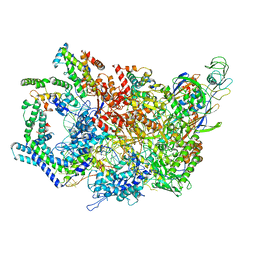 | | F. tularensis RNAPs70-(MglA-SspA)-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
7LQ4
 
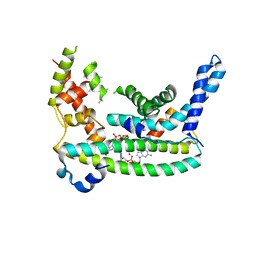 | | Rr (RsiG)2-(c-di-GMP)2-WhiG complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), RsiG, WhiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LQ2
 
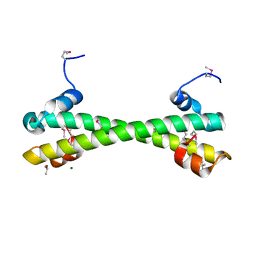 | | Apo Rr RsiG- crystal form 1 | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, RR RsiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1WET
 
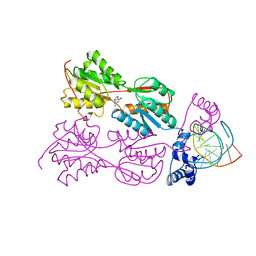 | | STRUCTURE OF THE PURR-GUANINE-PURF OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Glasfeld, A, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-04-27 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity.
J.Biol.Chem., 272, 1997
|
|
7LQ3
 
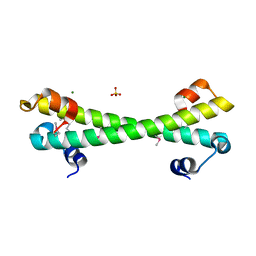 | |
3OQO
 
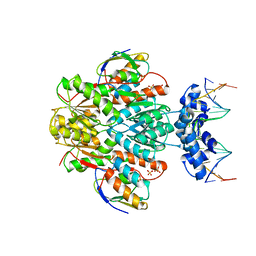 | | Ccpa-hpr-ser46p-syn cre | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Catabolite control protein A, ... | | Authors: | schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2011-10-26 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3OQN
 
 | | Structure of ccpa-hpr-ser46-p-gntr-down cre | | Descriptor: | 5'-D(*AP*TP*GP*GP*TP*AP*CP*CP*GP*CP*TP*TP*TP*CP*AP*A)-3', 5'-D(*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*TP*AP*CP*CP*AP*T)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3OQM
 
 | | structure of ccpa-hpr-ser46p-ackA2 complex | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*GP*TP*AP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*A)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
3IHQ
 
 | |
3BT9
 
 | |
3BTI
 
 | |
3BTC
 
 | |
2PUE
 
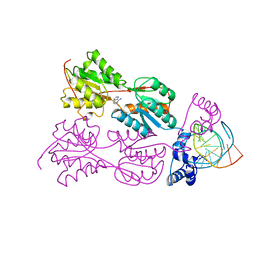 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | ADENINE, DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), PROTEIN (PURINE REPRESSOR ) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
1Z3E
 
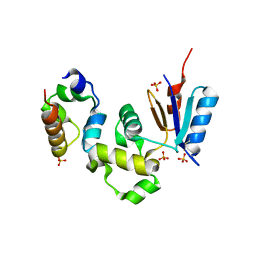 | | Crystal Structure of Spx in Complex with the C-terminal Domain of the RNA Polymerase Alpha Subunit | | Descriptor: | DNA-directed RNA polymerase alpha chain, Regulatory protein spx, SULFATE ION | | Authors: | Newberry, K.J, Nakano, S, Zuber, P, Brennan, R.G. | | Deposit date: | 2005-03-11 | | Release date: | 2005-10-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the {alpha} C-terminal domain of RNA polymerase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZAY
 
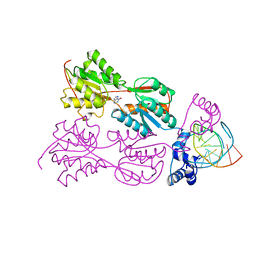 | | PURINE REPRESSOR-HYPOXANTHINE-MODIFIED-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*TP*(1AP)P*TP*TP*TP*TP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Zheleznova, E.E, Brennan, R.G. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Roles of Exocyclic Groups in the Central Base-Pair Step in Modulating the Affinity of PurR for its Operator
To be Published
|
|
1Z9C
 
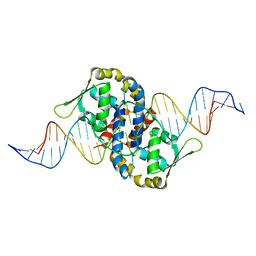 | | Crystal structure of OhrR bound to the ohrA promoter: Structure of MarR family protein with operator DNA | | Descriptor: | DNA (29-MER), Organic hydroperoxide resistance transcriptional regulator | | Authors: | Hong, M, Fuangthong, M, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2005-04-01 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family.
Mol.Cell, 20, 2005
|
|
1Z91
 
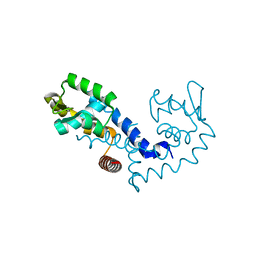 | |
1ZVV
 
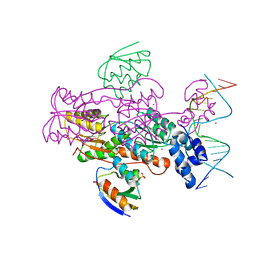 | | Crystal structure of a ccpa-crh-dna complex | | Descriptor: | DNA recognition strand CRE, Glucose-resistance amylase regulator, HPr-like protein crh, ... | | Authors: | Schumacher, M.A, Brennan, R.G, Hillen, W, Seidel, G. | | Deposit date: | 2005-06-02 | | Release date: | 2006-02-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J.Biol.Chem., 281, 2006
|
|
3D71
 
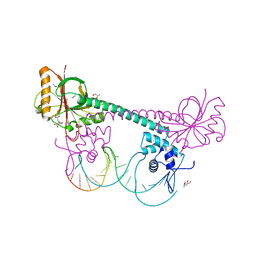 | | Crystal structure of E253Q BMRR bound to 22 base pair promoter site | | Descriptor: | BMR promoter DNA, CITRATE ANION, IMIDAZOLE, ... | | Authors: | Newberry, K.J, Huffman, J.L, Brennan, R.G. | | Deposit date: | 2008-05-20 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of BmrR-Drug Complexes Reveal a Rigid Multidrug Binding Pocket and Transcription Activation through Tyrosine Expulsion
J.Biol.Chem., 283, 2008
|
|
3IAO
 
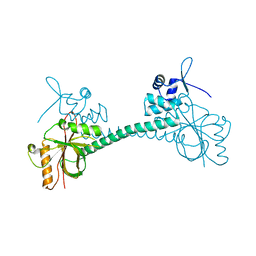 | |
1R8D
 
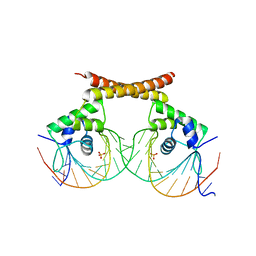 | | Crystal Structure of MtaN Bound to DNA | | Descriptor: | 26-MER, SULFATE ION, transcription activator MtaN | | Authors: | Newberry, K.J, Brennan, R.G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
J.Biol.Chem., 279, 2004
|
|
2BSQ
 
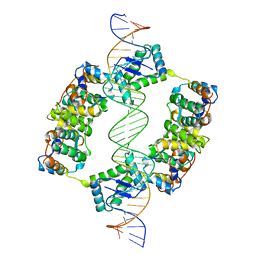 | | FitAB bound to DNA | | Descriptor: | IR36, FORWARD STRAND, REVERSE STRAND, ... | | Authors: | Mattison, K, Wilbur, J.S, So, M, Brennan, R.G. | | Deposit date: | 2005-05-23 | | Release date: | 2006-08-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Fitab from Neisseria Gonorrhoeae Bound to DNA Reveals a Tetramer of Toxin-Antitoxin Heterodimers Containing Pin Domains and Ribbon-Helix-Helix Motifs.
J.Biol.Chem., 281, 2006
|
|
2BOW
 
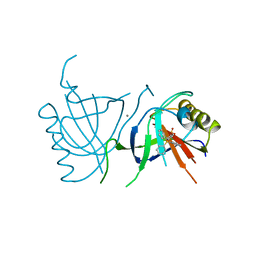 | | MULTIDRUG-BINDING DOMAIN OF TRANSCRIPTION ACTIVATOR BMRR IN COMPLEX WITH A LIGAND, TETRAPHENYLPHOSPHONIUM | | Descriptor: | MANGANESE (II) ION, MULTIDRUG-EFFLUX TRANSPORTER 1 REGULATOR BMRR, SULFATE ION, ... | | Authors: | Zheleznova, E.E, Markham, P.N, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter.
Cell(Cambridge,Mass.), 96, 1999
|
|
2EV0
 
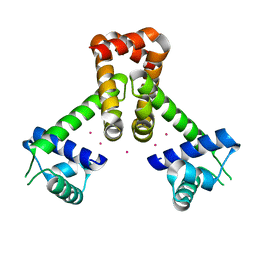 | | Bacillus subtilis manganese transport regulator (MNTR) bound to cadmium | | Descriptor: | CADMIUM ION, Transcriptional regulator mntR | | Authors: | Kliegman, J.I, Griner, S.L, Helmann, J.D, Brennan, R.G, Glasfeld, A. | | Deposit date: | 2005-10-30 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Metal-Selective Activation of the Manganese Transport Regulator of Bacillus subtilis.
Biochemistry, 45, 2006
|
|
2EV5
 
 | | Bacillus subtilis manganese transport regulator (MNTR) bound to calcium | | Descriptor: | CALCIUM ION, Transcriptional regulator mntR | | Authors: | Kliegman, J.I, Griner, S.L, Helmann, J.D, Brennan, R.G, Glasfeld, A. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Metal-Selective Activation of the Manganese Transport Regulator of Bacillus subtilis.
Biochemistry, 45, 2006
|
|
