3A2O
 
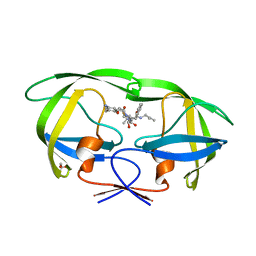 | | Crystal Structure of HIV-1 Protease Complexed with KNI-1689 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(4-amino-2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-5,5-dimethyl-N-(2-methylprop -2-en-1-yl)-1,3-thiazolidine-4-carboxamide, GLYCEROL, PROTEASE | | Authors: | Adachi, M, Tamada, T, Hidaka, K, Kimura, T, Kiso, Y, Kuroki, R. | | Deposit date: | 2009-05-26 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Small-sized human immunodeficiency virus type-1 protease inhibitors containing allophenylnorstatine to explore the S2' pocket.
J.Med.Chem., 52, 2009
|
|
1WDS
 
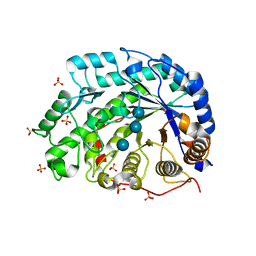 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
5X23
 
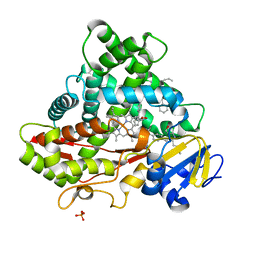 | | Crystal structure of CYP2C9 genetic variant A477T (*30) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5X24
 
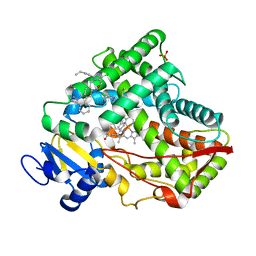 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | Descriptor: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Maekawa, K, Adachi, M, Shah, M.B. | | Deposit date: | 2017-01-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
6KIL
 
 | | N21Q mutant thioredoxin from Halobacterium salinarum NRC-1 | | Descriptor: | Thioredoxin | | Authors: | Arai, S, Shibazaki, C, Shimizu, R, Adachi, M, Ishibashi, M, Tokunaga, H, Tokunaga, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic mechanism and evolutional characteristics of thioredoxin from Halobacterium salinarum NRC-1.
Acta Crystallogr.,Sect.D, 76, 2020
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
5ZN4
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148N mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN0
 
 | | Joint X-ray/neutron structure of protein kinase ck2 alpha subunit | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Ostermann, A, Schrader, T.E, Sunami, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN1
 
 | | X-ray structure of protein kinase ck2 alpha subunit in D2O | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN5
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148A mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN2
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148A mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN3
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148S mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
1WDP
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDR
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDQ
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS1
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS5
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2B) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRZ
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS2
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1C) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1Q6C
 
 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6E
 
 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 5.4 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
