3MWM
 
 | |
4OOU
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
4OOZ
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose | | Descriptor: | Beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
1ZKJ
 
 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
4MB2
 
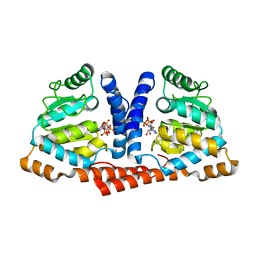 | | Crystal structure of TON1374 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphopantothenate synthetase | | Authors: | Kim, M.-K, An, Y.J, Cha, S.-S. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a novel phosphopantothenate synthetase from the hyperthermophilic archaea, Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 439, 2013
|
|
6K8X
 
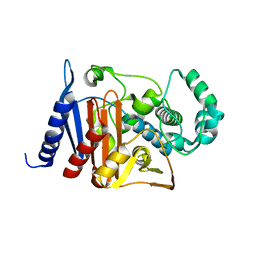 | | Crystal structure of a class C beta lactamase | | Descriptor: | Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KA5
 
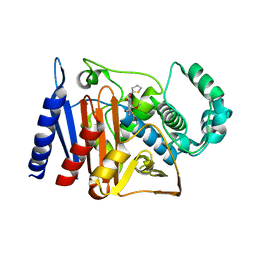 | | Crystal structure of a class C beta-lactamase in complex with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6K9T
 
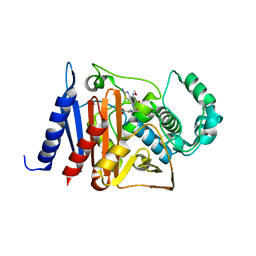 | | Crystal structure of a class C beta-lactamase in complex with cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KBY
 
 | | Crystal structure of a class C beta lactamase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
4FC5
 
 | | Crystal Structure of Ton_0340 | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, An, Y.J, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5IP5
 
 | |
4J4K
 
 | | Crystal structure of glucose isomerase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Xylose isomerase, ... | | Authors: | Kim, M.K, An, Y.J, Lee, S, Jeong, C.S, Cha, S.S. | | Deposit date: | 2013-02-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glucose isomerase
To be Published
|
|
5GGW
 
 | | Crystal structure of Class C beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3BWE
 
 | | Crystal structure of aggregated form of DJ1 | | Descriptor: | PHOSPHATE ION, Protein DJ-1 | | Authors: | Cha, S.S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of filamentous aggregates of human DJ-1 formed in an inorganic phosphate-dependent manner.
J.Biol.Chem., 283, 2008
|
|
2PQJ
 
 | | Crystal structure of active ribosome inactivating protein from maize (b-32), complex with adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Au, S.W.N, Cha, S.S, Young, J.A, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2PQI
 
 | | Crystal structure of active ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2PQG
 
 | | Crystal structure of inactive ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
5JOC
 
 | | Crystal structure of the S61A mutant of AmpC BER | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Na, J.H, An, Y.J, Cha, S.S. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4DWZ
 
 | | Crystal Structure of Ton_0340 | | Descriptor: | Hypothetical protein TON_0340, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5GL3
 
 | | Crystal structure of TON_0340 in complex with Mg | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL4
 
 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GKX
 
 | | Crystal structure of TON_0340, apo form | | Descriptor: | PHOSPHATE ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL2
 
 | | Crystal structure of TON_0340 in complex with Ca | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
