7X5D
 
 | |
7XRO
 
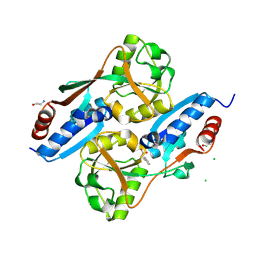 | |
5X0Q
 
 | | OxyR2 E204G variant (Cl-bound) from Vibrio vulnificus | | Descriptor: | CHLORIDE ION, CITRIC ACID, LysR family transcriptional regulator | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2017-01-23 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints
J. Biol. Chem., 292, 2017
|
|
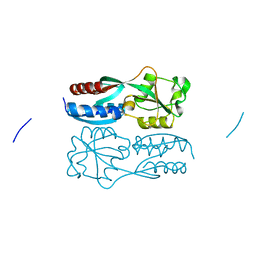
5X0N
 
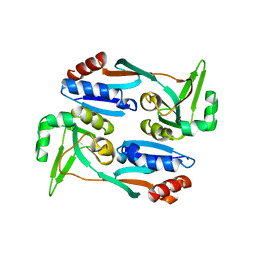 | |
5X0O
 
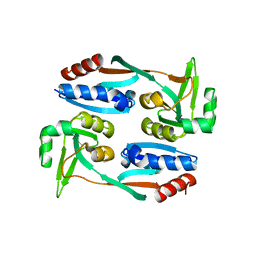 | |
5B7H
 
 | |
1HDU
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with aminocarbonylphenylalanine at 1.75 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(AMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HEE
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with L-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, L-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1HDQ
 
 | | Crystal structure of bovine pancreatic carboxypeptidase A complexed with D-N-hydroxyaminocarbonyl phenylalanine at 2.3 A | | Descriptor: | CARBOXYPEPTIDASE A, D-[(N-HYDROXYAMINO)CARBONYL]PHENYLALANINE, ZINC ION | | Authors: | Cho, J.H, Ha, N.-C, Chung, S.J, Kim, D.H, Choi, K.Y, Oh, B.-H. | | Deposit date: | 2000-11-17 | | Release date: | 2001-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into the Stereochemistry in the Inhibition of Carboxypeptidase a with N-(Hydroxyaminocarbonyl)Phenylalanine: Binding Modes of an Enantiomeric Pair of the Inhibitor to Carboxypeptidase A
Bioorg.Med.Chem., 10, 2002
|
|
1OHU
 
 | | Structure of Caenorhabditis elegans CED-9 | | Descriptor: | APOPTOSIS REGULATOR CED-9 | | Authors: | Jeong, J.-S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 2003-05-31 | | Release date: | 2003-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Unique Structural Features of a Bcl-2 Family Protein Ced-9 and Biophysical Characterization of Ced-9/Egl-1 Interactions
Cell Death Differ., 10, 2003
|
|
7DQG
 
 | |
7DSG
 
 | |
1EA2
 
 | | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Tyrosine-to-Phenylalanine Substitution(s) in the Hydrogen Bond Network of Delta-5-3-Ketosteroid Isomerase from Pheudomonas putida Biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Choi, G, Ha, N.-C, Kim, M.-S, Hong, B.-H, Choi, K.Y. | | Deposit date: | 2000-11-03 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudoreversion of the Catalytic Activity of Y14F by the Additional Substitution(S) of Tyrosine with Phenylalanine in the Hydrogen Bond Network of Delta (5)-3-Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochemistry, 40, 2001
|
|
2POO
 
 | | THERMOSTABLE PHYTASE IN FULLY CALCIUM LOADED STATE | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
4GQZ
 
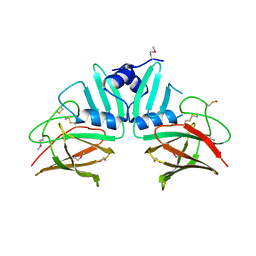 | | Crystal Structure of S.CueP | | Descriptor: | CHLORIDE ION, Putative periplasmic or exported protein | | Authors: | Ha, N.C, Yoon, B.Y. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-14 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure of the periplasmic copper-binding protein CueP from Salmonella enterica serovar Typhimurium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6KYY
 
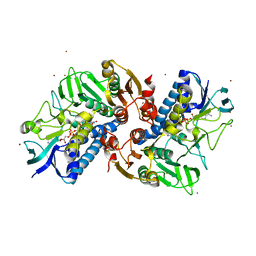 | |
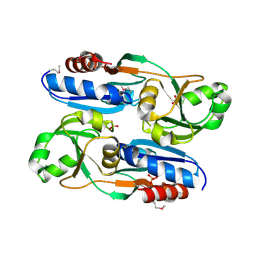
6L33
 
 | |
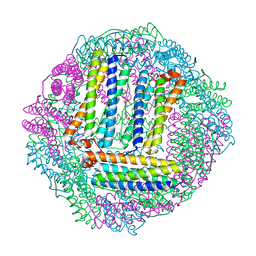
3GVY
 
 | |
3FTJ
 
 | |
6AKV
 
 | | Crystal structure of LysB4, the endolysin from Bacillus cereus-targeting bacteriophage B4 | | Descriptor: | CHLORIDE ION, LysB4, SULFATE ION, ... | | Authors: | Hong, S, Ha, N.-C. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of LysB4, an Endolysin fromBacillus cereus-Targeting Bacteriophage B4.
Mol. Cells, 42, 2019
|
|
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
2B9L
 
 | | Crystal structure of prophenoloxidase activating factor-II from the beetle Holotrichia diomphalia | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Piao, S, Song, Y.-L, Park, S.Y, Lee, B.L, Oh, B.-H, Ha, N.-C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a clip-domain serine protease and functional roles of the clip domains.
Embo J., 24, 2005
|
|
5K2J
 
 | |
5K2I
 
 | |
5K1G
 
 | | Crystal structure of reduced Prx3 from Vibrio vulnificus | | Descriptor: | 1-Cys peroxiredoxin | | Authors: | Ahn, J, Ha, N.-C. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of peroxiredoxin 3 fromVibrio vulnificusand its implications for scavenging peroxides and nitric oxide.
IUCrJ, 5, 2018
|
|
