1GMY
 
 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
6FJL
 
 | | Structure of IbpS from Dickeya dadantii | | Descriptor: | ABC-type Fe3+ transport system, periplasmic component, ACETATE ION, ... | | Authors: | Gueguen-Chaignon, V, Condemine, G, Terradot, L. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.70000041 Å) | | Cite: | A secreted metal-binding protein protects necrotrophic phytopathogens from reactive oxygen species.
Nat Commun, 10, 2019
|
|
2OA5
 
 | | Crystal structure of ORF52 from Murid herpesvirus (MUHV-4) (Murine gammaherpesvirus 68) at 2.1 A resolution. Northeast Structural Genomics Consortium target MHR28B. | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Hypothetical protein BQLF2 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Cunningham, K, Ma, L.-C, Ho, C.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68.
J.Biol.Chem., 282, 2007
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
2NW7
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferric heme. Northeast Structural Genomics Target XcR13 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Bruckmann, C, Thackray, S.J, Seetharaman, J, Tucker, T, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Chapman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
3QAE
 
 | | 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Streptococcus pneumoniae | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme a reductase, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhang, L, Feng, L, Zhou, L, Gui, J, Wan, J. | | Deposit date: | 2011-01-10 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase of Streptococcus pneumoniae
To be Published
|
|
3MX0
 
 | | Crystal Structure of EphA2 ectodomain in complex with ephrin-A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 2, ... | | Authors: | Himanen, J.P, Yermekbayeva, L, Janes, P.W, Walker, J.R, Xu, K, Atapattu, L, Rajashankar, K.R, Mensinga, A, Lackmann, M, Nikolov, D.B, Dhe-Paganon, S. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XN2
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose | | Descriptor: | ALPHA-GALACTOSIDASE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
1GIW
 
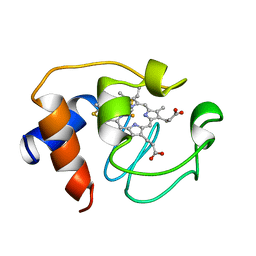 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-17 | | Release date: | 1998-12-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
1GKT
 
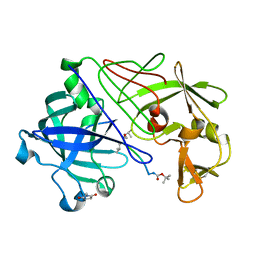 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
1GJP
 
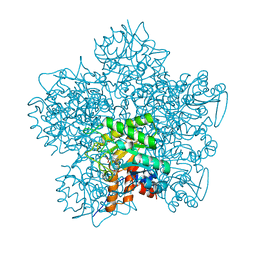 | | SCHIFF-BASE COMPLEX OF YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE WITH 4-OXOSEBACIC ACID | | Descriptor: | 4-OXODECANEDIOIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
3N0T
 
 | | Human dipeptidil peptidase DPP7 complexed with inhibitor GSK237826A | | Descriptor: | (3S)-4-oxo-4-piperidin-1-ylbutane-1,3-diamine, Dipeptidyl peptidase 2 | | Authors: | Dobrovetsky, E, Khutoreskaya, G, Seitova, A, Crombet, L, Cossar, D, Pagannon, S, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hassell, A, Shewchuk, L, Haffner, C, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-14 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Human dipeptidyl peptidase DPP7
To be Published
|
|
5XPV
 
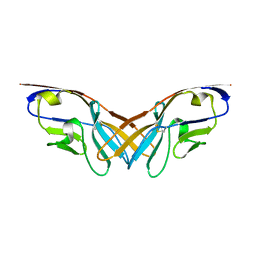 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
4JJY
 
 | | Alix V domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Pashkova, N, Gakhar, L, Yu, L, Piper, R.C. | | Deposit date: | 2013-03-08 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (6.503 Å) | | Cite: | The yeast alix homolog bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes.
Dev.Cell, 25, 2013
|
|
1GCB
 
 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
1FUQ
 
 | | FUMARASE WITH BOUND 3-TRIMETHYLSILYLSUCCINIC ACID | | Descriptor: | 3-TRIMETHYLSILYLSUCCINIC ACID, CITRIC ACID, FUMARASE C | | Authors: | Weaver, T, Banaszak, L. | | Deposit date: | 1996-08-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli.
Biochemistry, 35, 1996
|
|
6G30
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Stach, L, Morgan, R.M.L, Freemont, P.S. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
3N2B
 
 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
3N3W
 
 | | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni | | Descriptor: | Ribonuclease III | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Nuclease Domain of Ribonuclase III (rnc) from Campylobacter jejuni
TO BE PUBLISHED
|
|
7GDP
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NAU-LAT-8502cac5-2 (Mpro-x11186) | | Descriptor: | (3S)-5-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-2,3-dihydro-1-benzofuran-3-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB1
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with STE-KUL-2e0d2e88-2 (Mpro-x10150) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(4-acetylpiperazin-1-yl)ethyl]naphthalene-1-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.289 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GE3
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714a760b-3 (Mpro-x11317) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-amino-4-methylpyridin-3-yl)-2-(3-chlorophenyl)acetamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GBG
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-1 (Mpro-x10329) | | Descriptor: | (2S)-2-(3-chlorophenyl)-N-(5-methylpyridazin-4-yl)butanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GE7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ADA-UCB-6c2cb422-3 (Mpro-x11354) | | Descriptor: | 2-(4-methylpyridin-3-yl)-N-(1,2,3,4-tetrahydroisoquinolin-8-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
