4RAY
 
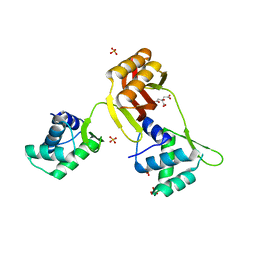 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Liu, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB1
 
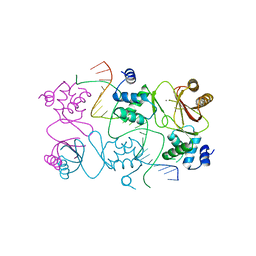 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Fur-Mn2+-E. coli Fur box | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*AP*TP*GP*AP*TP*AP*AP*TP*CP*AP*TP*TP*AP*TP*CP*CP*GP*C)-3'), DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB3
 
 | |
5IZ9
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
3K3O
 
 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
5IZ8
 
 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5IZ6
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, DI(HYDROXYETHYL)ETHER, PHQ-ALA-GLY-GLU-ALA-LEU-TYR-GLU-NH2, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
3K3N
 
 | | Crystal structure of the catalytic core domain of human PHF8 | | Descriptor: | FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
5IZA
 
 | | Protein-protein interaction | | Descriptor: | ACE-GLY-GLY-GLU-ALA-LEU-ALA-TRP-NH2, Adenomatous polyposis coli protein | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4ZHW
 
 | |
4ZHY
 
 | | Crystal structure of a bacterial signalling complex | | Descriptor: | FORMIC ACID, SULFATE ION, YfiB, ... | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHV
 
 | | Crystal structure of a bacterial signalling protein | | Descriptor: | SULFATE ION, YfiB | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
4ZHU
 
 | | Crystal structure of a bacterial repressor protein | | Descriptor: | SULFATE ION, YfiR | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.3968 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
5HE7
 
 | | BACE-1 in complex with (4aR,7aS)-7a-(2,4-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one | | Descriptor: | (2E,4aR,7aS)-7a-(2,4-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-16 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-Based Design of an Iminoheterocyclic beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE) Inhibitor that Lowers Central A beta in Nonhuman Primates.
J.Med.Chem., 59, 2016
|
|
5HDZ
 
 | |
5HE5
 
 | |
5I1V
 
 | |
5HDV
 
 | |
5I1W
 
 | | Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis | | Descriptor: | 4-hydroxy[2,2'-bipyridine]-6-carbaldehyde, 6-(hydroxymethyl)[2,2'-bipyridin]-4-ol, CrmK, ... | | Authors: | Picard, M.-E, Barma, J, Shi, R. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into flavoenzyme CrmK reveals a shunt product recycling mechanism in caerulomycin biosynthesis
to be published
|
|
5HD0
 
 | |
5HDU
 
 | |
5HDX
 
 | |
5HE4
 
 | | BACE-1 in complex with (4aR,7aS)-7a-(2,6-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-3-methyl-4-oxooctahydro-2H-pyrrolo[3,4-d]pyrimidin-2-iminium | | Descriptor: | (2E,4aR,7aS)-7a-(2,6-difluorophenyl)-6-(5-fluoro-4-methoxy-6-methylpyrimidin-2-yl)-2-imino-3-methyloctahydro-4H-pyrrolo[3,4-d]pyrimidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Orth, P. | | Deposit date: | 2016-01-05 | | Release date: | 2016-03-16 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of an Iminoheterocyclic beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE) Inhibitor that Lowers Central A beta in Nonhuman Primates.
J.Med.Chem., 59, 2016
|
|
3LPI
 
 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
