3ESF
 
 | |
6T7P
 
 | | human plasmakallikrein protease domain in complex with active site directed inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-6-yl]carbonyl]-~{N}-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Renatus, M. | | Deposit date: | 2019-10-22 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TS5
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-propan-2-yl-phenyl]ethoxy]-3-methoxy-benzoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
1DY6
 
 | | Structure of the imipenem-hydrolyzing beta-lactamase SME-1 | | Descriptor: | CARBAPENEM-HYDROLYSING BETA-LACTAMASE SME-1 | | Authors: | Sougakoff, W, L'Hermite, G, Billy, I, Guillet, V, Naas, T, Nordman, P, Jarlier, V, Delettre, J. | | Deposit date: | 2000-01-27 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the Imipenem-Hydrolyzing Class a Beta-Lactamase Sme-1 from Serratia Marcescens.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6TS6
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-(2-cyanopropan-2-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TS4
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TS7
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-(1,2,3,4-tetrahydroisoquinolin-7-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6GYH
 
 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
4S2M
 
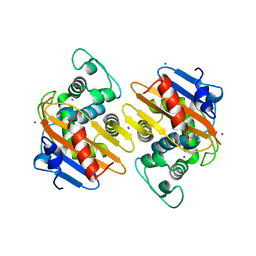 | | Crystal Structure of OXA-163 complexed with iodide in the active site | | Descriptor: | Beta-lactamase, IODIDE ION | | Authors: | Stojanoski, V, Hu, L, Palzkill, T.G, Prasad, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
7ZHR
 
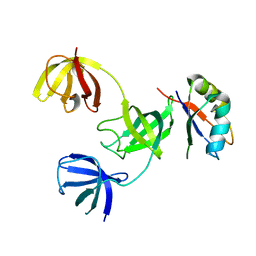 | |
7ZHH
 
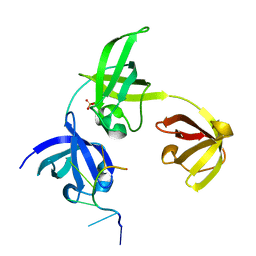 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
4S2L
 
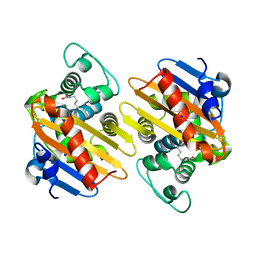 | | Crystal Structure of OXA-163 beta-lactamase | | Descriptor: | Beta-lactamase, SODIUM ION | | Authors: | Stojanoski, V, Liya, H, Palzkill, T.G, Prasad, B, Sankaran, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
4RMW
 
 | |
4RMU
 
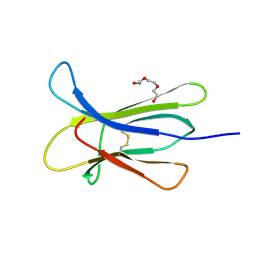 | |
4RMV
 
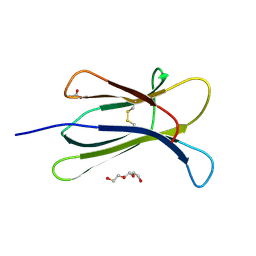 | |
3NOL
 
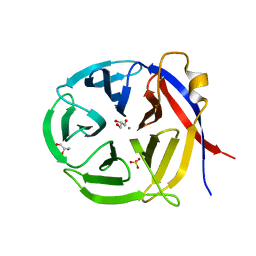 | | Crystal structure of Zymomonas mobilis Glutaminyl Cyclase (trigonal form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3NOK
 
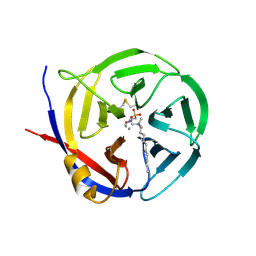 | | Crystal structure of Myxococcus xanthus Glutaminyl Cyclase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
1GFF
 
 | |
2PUO
 
 | |
2PVD
 
 | | Crystal srtucture of the reduced ferredoxin:thioredoxin reductase | | Descriptor: | Ferredoxin-thioredoxin reductase, catalytic chain, variable chain, ... | | Authors: | Dai, S. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural snapshots along the reaction pathway of ferredoxin-thioredoxin reductase.
Nature, 448, 2007
|
|
2PVG
 
 | |
2PVO
 
 | |
2PU9
 
 | |
1AOK
 
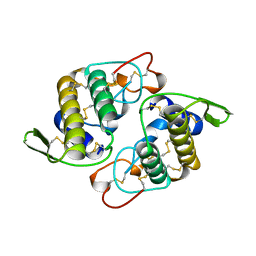 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
5NAJ
 
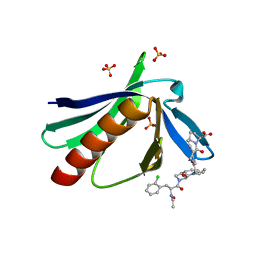 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-1]-[ProM-1]-OH | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{R},7~{S},10~{S},13~{R})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{S},7~{R},10~{R},13~{S})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, CHLORIDE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
