7K79
 
 | | CBF3 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K7G
 
 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K78
 
 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7D2T
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Yang, H, Wei, Z, Yu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7VAH
 
 | |
3HDN
 
 | |
7D2S
 
 | | Crystal structure of Rsu1/PINCH1_LIM5C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, Ras suppressor protein 1, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7D2U
 
 | | Crystal structure of Rsu1/PINCH1_LIM45C complex | | Descriptor: | GLYCEROL, LIM and senescent cell antigen-like-containing domain protein 1, MALONATE ION, ... | | Authors: | Yang, H, Wei, Z, Cong, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics.
Elife, 10, 2021
|
|
7F83
 
 | | Crystal Structure of a receptor in Complex with inverse agonist | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-methylimidazo[2,1-b][1,3]thiazol-6-yl)-1-[2-[(1R)-5-(6-methylpyrimidin-4-yl)-2,3-dihydro-1H-inden-1-yl]-2,7-diazaspiro[3.5]nonan-7-yl]ethanone, Growth hormone secretagogue receptor type 1,Soluble cytochrome b562 | | Authors: | Xu, Z, Shao, Z. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
3HDM
 
 | |
1D4T
 
 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP IN COMPLEX WITH A SLAM PEPTIDE | | Descriptor: | SIGNALING LYMPHOCYTIC ACTIVATION MOLECULE, T CELL SIGNAL TRANSDUCTION MOLECULE SAP | | Authors: | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
5GXJ
 
 | | Zika Virus NS2B-NS3 protease | | Descriptor: | FLAVIVIRUS_NS2B,LINKER,Peptidase S7 | | Authors: | Yang, H, Chen, X, Ji, X, Xiong, Y, Yang, K. | | Deposit date: | 2016-09-18 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Viral protease activation mechanism
To Be Published
|
|
7W2Z
 
 | | Cryo-EM structure of the ghrelin-bound human ghrelin receptor-Go complex | | Descriptor: | Appetite-regulating hormone, CHOLESTEROL, Growth hormone secretagogue receptor type 1, ... | | Authors: | Qin, J, Ming, Q, Ji, S, Mao, C, Shen, D, Zhang, Y. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of agonism and inverse agonism in ghrelin receptor.
Nat Commun, 13, 2022
|
|
5V8C
 
 | | LytR-Csp2A-Psr enzyme from Actinomyces oris | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Amer, B.R, Sawaya, M.R, Liauw, B, Fu, J.Y, Ton-That, H, Clubb, R.T. | | Deposit date: | 2017-03-21 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein.
MBio, 10, 2019
|
|
5VAO
 
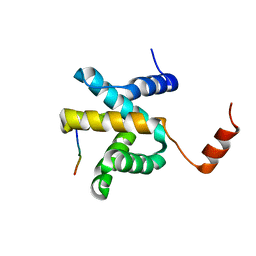 | | Crystal structure of eVP30 C-terminus and eNP peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | XU, W, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis.
Nat Commun, 8, 2017
|
|
1D1Z
 
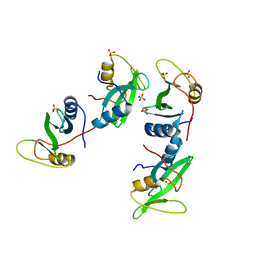 | | CRYSTAL STRUCTURE OF THE XLP PROTEIN SAP | | Descriptor: | SAP SH2 DOMAIN, SULFATE ION | | Authors: | Poy, F, Yaffe, M.B, Sayos, J, Saxena, K, Eck, M.J. | | Deposit date: | 1999-09-22 | | Release date: | 1999-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition.
Mol.Cell, 4, 1999
|
|
8GVK
 
 | |
8GZZ
 
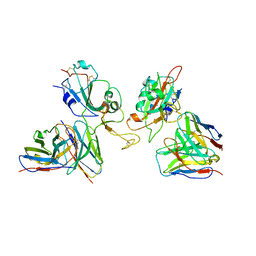 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | Descriptor: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8H01
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8H00
 
 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8HEC
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8H0F
 
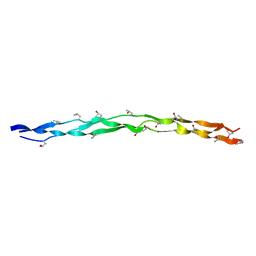 | | Crystal structure of collagen heterotrimer with KD,ER and KE axial pairs | | Descriptor: | collagen-like peptide chain A, collagen-like peptide chain B, collagen-like peptide chain C | | Authors: | Fan, S. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Biophys.J., 122, 2023
|
|
8GZO
 
 | |
