7K0S
 
 | |
2HJV
 
 | |
3II5
 
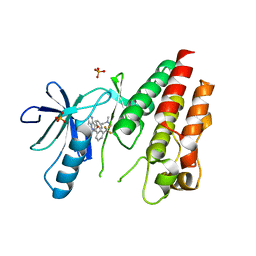 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IND
 
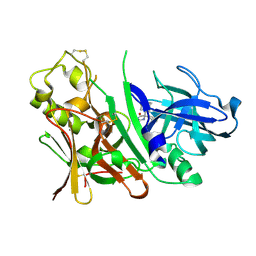 | | Bace1 with the aminohydantoin Compound 29 | | Descriptor: | (5S)-2-amino-3-methyl-5-phenyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-08-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Design and synthesis of 5,5'-disubstituted aminohydantoins as potent and selective human beta-secretase (BACE1) inhibitors.
J.Med.Chem., 53, 2010
|
|
3INE
 
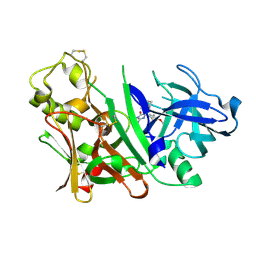 | | Bace1 with the aminohydantoin Compound S-34 | | Descriptor: | (5S)-2-amino-5-(4-methoxy-3-methylphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-08-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Design and synthesis of 5,5'-disubstituted aminohydantoins as potent and selective human beta-secretase (BACE1) inhibitors.
J.Med.Chem., 53, 2010
|
|
3IOP
 
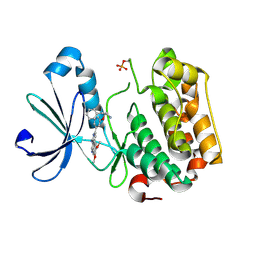 | | PDK-1 in complex with the inhibitor Compound-8i | | Descriptor: | 2-(5-{[(2R)-2-amino-3-phenylpropyl]oxy}pyridin-3-yl)-8,9-dimethoxybenzo[c][2,7]naphthyridin-4-amine, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Eur.J.Med.Chem., 45, 2010
|
|
3INH
 
 | | Bace1 with the aminohydantoin Compound R-58 | | Descriptor: | (5R)-2-amino-5-(4-fluoro-3-pyrimidin-5-ylphenyl)-3-methyl-5-[4-(trifluoromethoxy)phenyl]-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-08-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of 5,5'-disubstituted aminohydantoins as potent and selective human beta-secretase (BACE1) inhibitors.
J.Med.Chem., 53, 2010
|
|
3ION
 
 | | PDK1 in complex with Compound 8h | | Descriptor: | 2-(5-{[(2S)-2-amino-3-phenylpropyl]oxy}pyridin-3-yl)-8,9-dimethoxybenzo[c][2,7]naphthyridin-4-amine, 3-phosphoinositide-dependent protein kinase 1, SULFATE ION | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Eur.J.Med.Chem., 45, 2010
|
|
3INF
 
 | | Bace1 with the aminohydantoin Compound 37 | | Descriptor: | (5S)-2-amino-5-(4-methoxy-3-methylphenyl)-3-methyl-5-(3-pyridin-3-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M, Chopra, R. | | Deposit date: | 2009-08-12 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Design and synthesis of 5,5'-disubstituted aminohydantoins as potent and selective human beta-secretase (BACE1) inhibitors.
J.Med.Chem., 53, 2010
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
4XDZ
 
 | | Holo structure of ketol-acid reductoisomerase from Ignisphaera aggregans | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-12-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
4XDY
 
 | | Structure of NADH-preferring ketol-acid reductoisomerase from an uncultured archean | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-12-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
4XEH
 
 | |
6IJ1
 
 | | Crystal structure of a protein from Actinoplanes | | Descriptor: | ACETATE ION, IMIDAZOLE, Prenylcyclase | | Authors: | Yang, Z.Z, Zhang, L.L, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-10-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Crystal structure of TchmY from Actinoplanes teichomyceticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5ZVV
 
 | | Structure of SeMet-phAimR | | Descriptor: | AimR transcriptional regulator, GLYCEROL | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW6
 
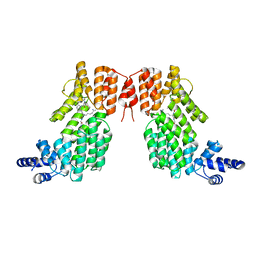 | | Structure of spAimR | | Descriptor: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW5
 
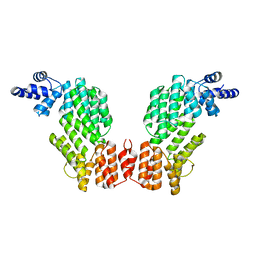 | | Structure of SeMet-spAimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-29 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVW
 
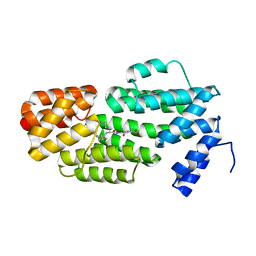 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZXL
 
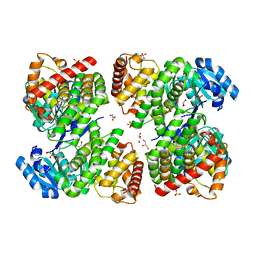 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
3QW9
 
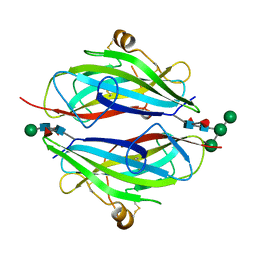 | | Crystal structure of betaglycan ZP-C domain | | Descriptor: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lin, S.J, Jardetzky, T.S. | | Deposit date: | 2011-02-27 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4JDZ
 
 | |
4JE0
 
 | |
4L7F
 
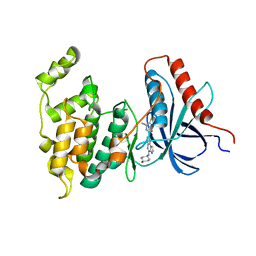 | | Co-crystal Structure of JNK1 and AX13587 | | Descriptor: | Mitogen-activated protein kinase 8, N-[1-(4-fluorophenyl)cyclopropyl]-4-[(trans-4-hydroxycyclohexyl)amino]imidazo[1,2-a]quinoxaline-8-carboxamide | | Authors: | Walter, R.L, Ranieri, G.M, Riggs, A.M, Weissig, H, Li, B, Shreder, K.R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hit-to-lead optimization and kinase selectivity of imidazo[1,2-a]quinoxalin-4-amine derived JNK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
