8HQM
 
 | | Activation mechanism of GPR132 by NPGLY | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
8H2F
 
 | |
8HI1
 
 | | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, DNA (26-MER), DNA (31-MER), ... | | Authors: | Chen, Q, Luo, Y. | | Deposit date: | 2022-11-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism.
Innovation (N Y), 4, 2023
|
|
8H18
 
 | |
2JSF
 
 | |
5ZOE
 
 | | Crystal Structure of D181A hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5GP1
 
 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5ZOD
 
 | | Crystal Structure of hFen1 in apo form | | Descriptor: | Flap endonuclease 1, MAGNESIUM ION, POTASSIUM ION | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5GOZ
 
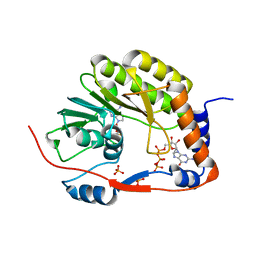 | |
5ZOG
 
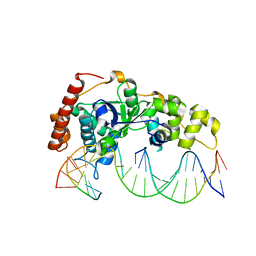 | | Crystal Structure of R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5WUA
 
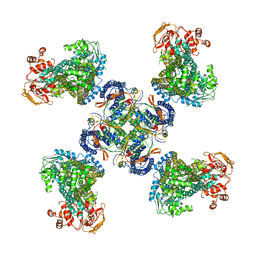 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
1F29
 
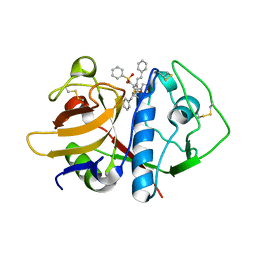 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (I) | | Descriptor: | 3-[[N-[MORPHOLIN-N-YL]-CARBONYL]-PHENYLALANINYL-AMINO]-5- PHENYL-PENTANE-1-SULFONYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F2A
 
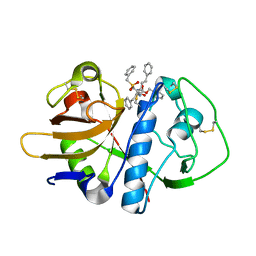 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO A VINYL SULFONE DERIVED INHIBITOR (II) | | Descriptor: | 3-[N-[BENZYLOXYCARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONYLMETHYLBENZENE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F2B
 
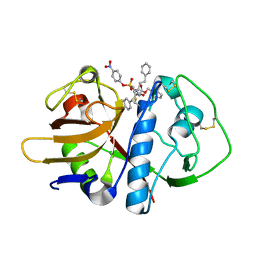 | | CRYSTAL STRUCTURE ANALYSIS OF CRUZAIN BOUND TO VINYL SULFONE DERIVED INHIBITOR (III) | | Descriptor: | 3-[N-[BENZYLOXYCARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID 4-NITRO-PHENYL ESTER, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
1F2C
 
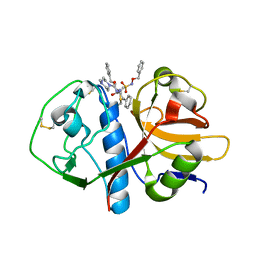 | | CRYSTAL STRUCTURE ANALYSIS OF CRYZAIN BOUND TO VINYL SULFONE DERIVED INHIBITOR (IV) | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, CRUZAIN | | Authors: | Brinen, L.S, Hansell, E, Roush, W.R, McKerrow, J.H, Fletterick, R.J. | | Deposit date: | 2000-05-23 | | Release date: | 2000-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A target within the target: probing cruzain's P1' site to define structural determinants for the Chagas' disease protease.
Structure Fold.Des., 8, 2000
|
|
7PL7
 
 | |
7CR6
 
 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CR8
 
 | | Synechocystis Cas1-Cas2-prespacerL complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
6ZVI
 
 | | Mbf1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S10-A, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
7DXJ
 
 | |
7DXK
 
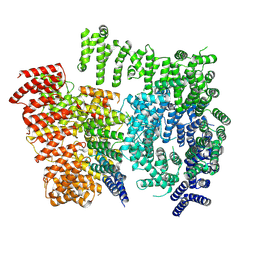 | |
6ZVH
 
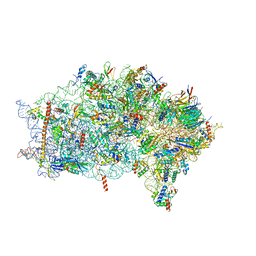 | | EDF1-ribosome complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Best, K.M, Denk, T, Cheng, J, Thoms, M, Berninghausen, O, Beckmann, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | EDF1 coordinates cellular responses to ribosome collisions.
Elife, 9, 2020
|
|
7X6W
 
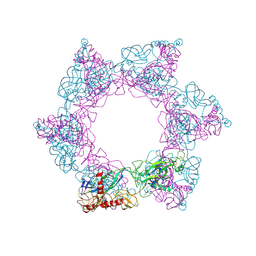 | | SFTSV 2 fold hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Z, Lou, Z. | | Deposit date: | 2022-03-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (5.18 Å) | | Cite: | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
7X6U
 
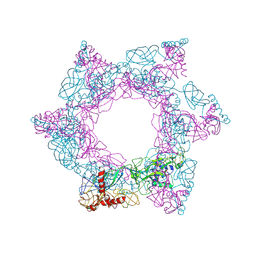 | | SFTSV 3 fold hexmer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Z, Lou, Z. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
7X72
 
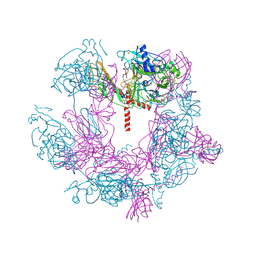 | | SFTSV 5 fold pentamer | | Descriptor: | Envelopment polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Z, Lou, Z. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Architecture of severe fever with thrombocytopenia syndrome virus.
Protein Cell, 14, 2023
|
|
