6TGS
 
 | | AtNBR1-PB1 domain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Jakobi, A.J, Sachse, C. | | Deposit date: | 2019-11-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake.
Nat Commun, 11, 2020
|
|
6TGN
 
 | |
1AEY
 
 | |
6TGP
 
 | |
6TGY
 
 | |
2FGO
 
 | | Structure of the 2[4FE-4S] ferredoxin from Pseudomonas aeruginosa | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER | | Authors: | Giastas, P, Pinotsis, N, Mavridis, I.M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure of the 2[4Fe-4S] ferredoxin from Pseudomonas aeruginosa at 1.32-A resolution: comparison with other high-resolution structures of ferredoxins and contributing structural features to reduction potential values.
J.Biol.Inorg.Chem., 11, 2006
|
|
2ILL
 
 | | Anomalous substructure of Titin-A168169 | | Descriptor: | CHLORIDE ION, Titin | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-10-03 | | Release date: | 2007-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2K0J
 
 | | Solution structure of CaM complexed to DRP1p | | Descriptor: | CALCIUM ION, LANTHANUM (III) ION, calmodulin | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-02-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
2K61
 
 | | Solution structure of CaM complexed to DAPk peptide | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J. | | Deposit date: | 2008-07-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
2G4L
 
 | | Anomalous substructure of hydroxynitrile lyase | | Descriptor: | (S)-acetone-cyanohydrin lyase, CHLORIDE ION, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4N
 
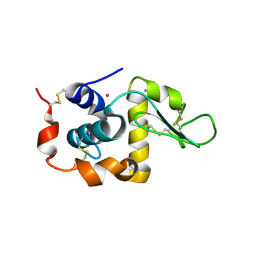 | | Anomalous substructure of alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, CALCIUM ION, POTASSIUM ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4Y
 
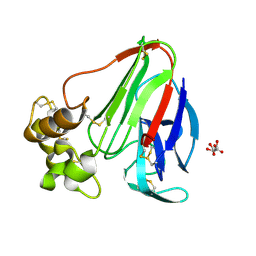 | | structure of thaumatin at 2.0 A wavelength | | Descriptor: | D(-)-TARTARIC ACID, Thaumatin-1 | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4J
 
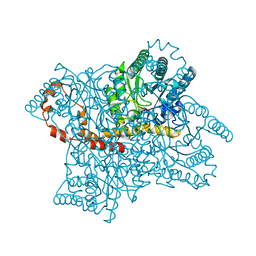 | | Anomalous substructure of Glucose isomerase | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4Q
 
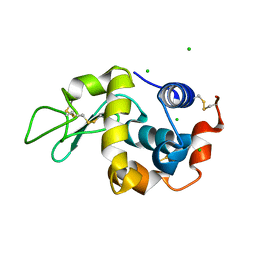 | | Anomalous substructure of lysozyme at pH 8.0 | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2G4Z
 
 | | anomalous substructure of thermolysin | | Descriptor: | CALCIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1H5Y
 
 | | HisF protein from Pyrobaculum aerophilum | | Descriptor: | GLYCEROL, HISF, PHOSPHATE ION | | Authors: | Banfield, M.J, Lott, J.S, McCarthy, A.A, Baker, E.N. | | Deposit date: | 2001-05-31 | | Release date: | 2001-06-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Hisf, a Histidine Biosynthetic Protein from Pyrobaculum Aerophilum
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3TQ5
 
 | |
3TS6
 
 | |
3TTA
 
 | |
3TSL
 
 | |
3TQ3
 
 | |
3TRN
 
 | |
3TPS
 
 | | Crystal structure of M-PMV dUTPASE complexed with dUPNPP substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
To be Published
|
|
3TPY
 
 | | Crystal structure of M-PMV dUTPase with a mixed population of substrate (dUPNPP) and post-inversion product (dUMP) in the active sites | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
to be published
|
|
3TP1
 
 | |
