2ZLY
 
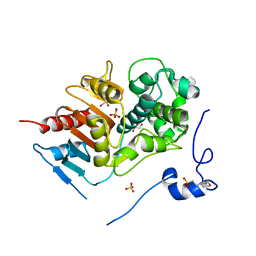 | | Structure of 6-aminohexanoate-dimer hydrolase, D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM9
 
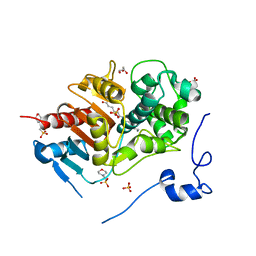 | | Structure of 6-Aminohexanoate-dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2DPT
 
 | |
2DY8
 
 | |
2E8J
 
 | |
2DY7
 
 | |
2CZY
 
 | | Solution structure of the NRSF/REST-mSin3B PAH1 complex | | Descriptor: | Paired amphipathic helix protein Sin3b, transcription factor REST (version 3) | | Authors: | Nomura, M, Uda-Tochio, H, Murai, K, Mori, N, Nishimura, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Neural Repressor NRSF/REST Binds the PAH1 Domain of the Sin3 Corepressor by Using its Distinct Short Hydrophobic Helix
J.Mol.Biol., 354, 2005
|
|
2DPS
 
 | |
2ZA6
 
 | | recombinant horse L-chain apoferritin | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Yamashita, I, Mishima, Y, Park, S.-Y, Heddle, J.G, Tame, J.R.H. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effect of N-terminal Residues on the Structural Stability of Recombinant Horse L-chain Apoferritin in an Acidic Environment
J.BIOCHEM.(TOKYO), 142, 2007
|
|
2ZA7
 
 | | recombinant horse L-chain apoferritin N-terminal deletion mutant (residues 1-4) | | Descriptor: | Ferritin light chain | | Authors: | Yamashita, I, Mishima, Y, Park, S.-Y, Heddle, J.G, Tame, J.R.H. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of N-terminal Residues on the Structural Stability of Recombinant Horse L-chain Apoferritin in an Acidic Environment
J.BIOCHEM.(TOKYO), 142, 2007
|
|
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
3APM
 
 | | Crystal structure of the human SNP PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZCZ
 
 | | Crystal structures and thermostability of mutant TRAP3 A7 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
3APN
 
 | | Crystal structure of the human wild-type PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Z33
 
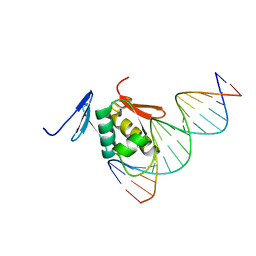 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | Descriptor: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
2ZA8
 
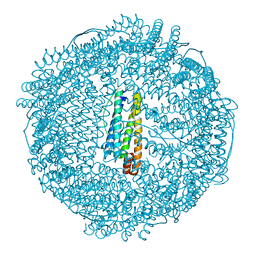 | | recombinant horse L-chain apoferritin N-terminal deletion mutant (residues 1-8) | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Yamashita, I, Mishima, Y, Park, S.-Y, Heddle, J.G, Tame, J.R.H. | | Deposit date: | 2007-10-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of N-terminal Residues on the Structural Stability of Recombinant Horse L-chain Apoferritin in an Acidic Environment
J.BIOCHEM.(TOKYO), 142, 2007
|
|
2ZD0
 
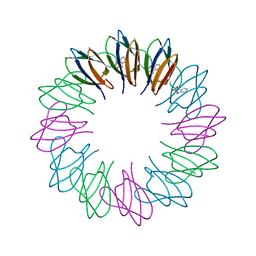 | | Crystal structures and thermostability of mutant TRAP3 A5 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
6JZH
 
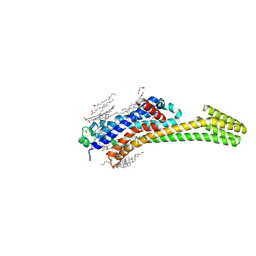 | | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Nango, E, Shimamura, T, Nakane, T, Yamanaka, Y, Mori, C, Kimura, K.T, Fujiwara, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
6JZI
 
 | | Structure of hen egg-white lysozyme obtained from SFX experiments under atmospheric pressure | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Nango, E, Sugahara, M, Nakane, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
2E8I
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
2ZMA
 
 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
7KZE
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, Leukotriene A-4 hydrolase, TRIETHYLENE GLYCOL, ... | | Authors: | Lee, K.H, Shim, Y, Paige, M, Noble, S.M. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
4YIR
 
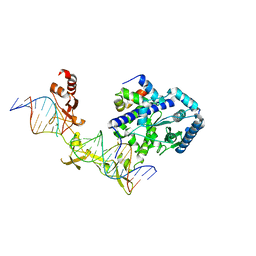 | | Crystal structure of Rad4-Rad23 crosslinked to an undamaged DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*G*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*GP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Min, J.-H, Chen, X, Kim, Y. | | Deposit date: | 2015-03-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0501 Å) | | Cite: | Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.
Nat Commun, 6, 2015
|
|
6UBF
 
 | |
7M2U
 
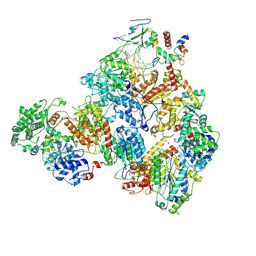 | | Nucleotide Excision Repair complex TFIIH Rad4-33 | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Murakami, K. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
