2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
2W1P
 
 | | 1.4 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 8.0 | | Descriptor: | AQUAPORIN PIP2-7 7;, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-20 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism
Plos Biol., 7, 2009
|
|
6FKR
 
 | | Crystal structure of the dolphin proline-rich antimicrobial peptide Tur1A bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16 ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Perebaskine, N, Benincasa, M, Gambato, S, Hofmann, S, Huter, P, Muller, C, Hilpert, K, Innis, C.A, Tossi, A, Wilson, D.N. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Dolphin Proline-Rich Antimicrobial Peptide Tur1A Inhibits Protein Synthesis by Targeting the Bacterial Ribosome.
Cell Chem Biol, 25, 2018
|
|
6G9E
 
 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis - TsES1 - 6 molecules in ASU | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
4A0O
 
 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
2UYA
 
 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
8OOR
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOK
 
 | |
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | Descriptor: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-04 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOC
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
6G9C
 
 | | Crystal structure of immunomodulatory active chitinase from Trichuris suis, TsES1 | | Descriptor: | 1,2-ETHANEDIOL, Immunomodulatory active chitinase | | Authors: | Malecki, P.H, Balster, K, Hartmann, S, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Helminth-Derived Chitinase Structurally Similar to Mammalian Chitinase Displays Immunomodulatory Properties in Inflammatory Lung Disease.
J Immunol Res, 2021, 2021
|
|
4A0W
 
 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.9 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A13
 
 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
2W2E
 
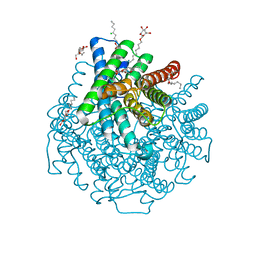 | | 1.15 Angstrom crystal structure of P.pastoris aquaporin, Aqy1, in a closed conformation at pH 3.5 | | Descriptor: | AQUAPORIN PIP2-7 7, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B.L, Neutze, R, Lindkvist-Petersson, K. | | Deposit date: | 2008-10-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Plos Biol., 7, 2009
|
|
4A0V
 
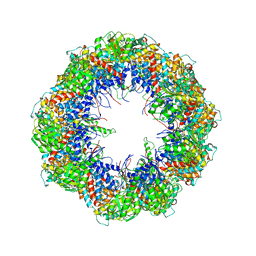 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
6U5O
 
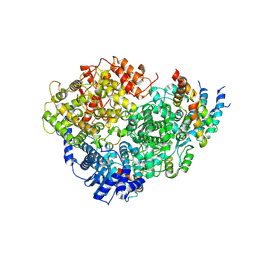 | | Structure of the Human Metapneumovirus Polymerase bound to the phosphoprotein tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Pan, J, Qian, X, Lattmann, S, Sahili, A.E, Yeo, T.H, Kalocsay, M, Fearns, R, Lescar, J. | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the human metapneumovirus polymerase phosphoprotein complex.
Nature, 577, 2020
|
|
4CN6
 
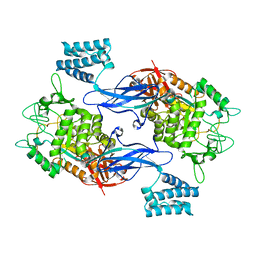 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with maltose bound | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
7MLH
 
 | | Crystal structure of human IgE (2F10) in complex with Der p 2.0103 | | Descriptor: | CHLORIDE ION, Der p 2 variant 3, IgE Heavy chain, ... | | Authors: | Khatri, K, Kapingidza, A.B, Richardson, C.M, Vailes, L.D, Wunschmann, S, Dolamore, C, Chapman, M.D, Smith, S.A, Pomes, A, Chruszcz, M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human IgE monoclonal antibody recognition of mite allergen Der p 2 defines structural basis of an epitope for IgE cross-linking and anaphylaxis in vivo.
Pnas Nexus, 1, 2022
|
|
8POI
 
 | |
8PX5
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
