6NZF
 
 | |
6NWZ
 
 | | Crystal structure of Agd3 a novel carbohydrate deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation.
Nat Commun, 11, 2020
|
|
7K7T
 
 | | Crystal structure of human MORC4 ATPase-CW in complex with AMPPNP | | Descriptor: | Isoform 3 of MORC family CW-type zinc finger protein 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular mechanism of the MORC4 ATPase activation.
Nat Commun, 11, 2020
|
|
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
6OQ5
 
 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3Q1T
 
 | |
5BTW
 
 | | Structure of the N-terminal domain of lpg1496 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Wong, K, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.
J.Biol.Chem., 290, 2015
|
|
6NZR
 
 | |
6NZE
 
 | |
6NZP
 
 | |
5USJ
 
 | | Crystal Structure of human KRAS G12D mutant in complex with GDPNP | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Huang, C.S, Kaplan, A, Stockwell, B.R, Tong, L. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Multivalent Small-Molecule Pan-RAS Inhibitors.
Cell, 168, 2017
|
|
3PA3
 
 | | X-ray crystal structure of compound 70 bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[2,3-d]pyridazine-7-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5T8Q
 
 | |
5TDW
 
 | | Set3 PHD finger in complex with histone H3K4me3 | | Descriptor: | SET domain-containing protein 3, SODIUM ION, ZINC ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-10-19 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
3OKS
 
 | |
6O6L
 
 | | The Structure of EgtB(Cabther) in complex with Hercynine | | Descriptor: | EgtB (Cabther), FE (III) ION, N,N,N-trimethyl-histidine | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
5DJO
 
 | | Crystal structure of the CC1-FHA tandem of Kinesin-3 KIF13A | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Ren, J.Q, Li, W, Huo, L, Feng, W. | | Deposit date: | 2015-09-02 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
J.Biol.Chem., 291, 2016
|
|
5SVI
 
 | |
5SVY
 
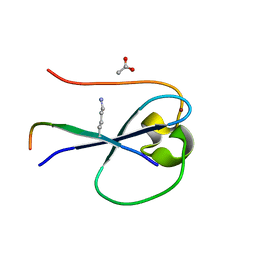 | | MORC3 CW in complex with histone H3K4me1 | | Descriptor: | ACETATE ION, H3K4me1, MORC family CW-type zinc finger protein 3, ... | | Authors: | Tong, Q, Andrews, F.H, Kutateladze, T.G. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Multivalent Chromatin Engagement and Inter-domain Crosstalk Regulate MORC3 ATPase.
Cell Rep, 16, 2016
|
|
3PPI
 
 | |
3LLS
 
 | |
6OJB
 
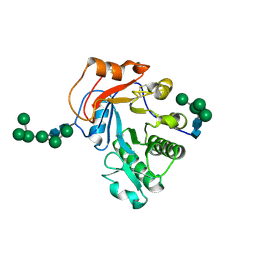 | | Crystal Structure of Aspergillus fumigatus Ega3 complex with galactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Bamford, N.C, Howell, P.L. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
6OQ6
 
 | |
6O6M
 
 | | The Structure of EgtB (Cabther) | | Descriptor: | EgtB (Cabther), FE (III) ION, GLYCEROL | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
3LGZ
 
 | | Crystal structure of dehydrosqualene synthase Y129A from S. aureus complexed with presqualene pyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Lin, F.-Y, Liu, Y.-L, Liu, C.-I, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-21 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
