2RRH
 
 | | NMR structure of vasoactive intestinal peptide in Methanol | | Descriptor: | VIP peptides | | Authors: | Umetsu, Y, Tenno, T, Goda, N, Ikegami, T, Hiroaki, H. | | Deposit date: | 2010-11-12 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural difference of vasoactive intestinal peptide in two distinct membrane-mimicking environments.
Biochim.Biophys.Acta, 1814, 2011
|
|
3QBI
 
 | |
3QBG
 
 | | Anion-free blue form of pharaonis halorhodopsin | | Descriptor: | BACTERIORUBERIN, Halorhodopsin, RETINAL, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an O-like blue form and an anion-free yellow form of pharaonis halorhodopsin
J.Mol.Biol., 413, 2011
|
|
1VSH
 
 | | ASV INTEGRASE CORE DOMAIN WITH ZN(II) COFACTORS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE, ZINC ION | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity.
J.Biol.Chem., 272, 1997
|
|
3VVK
 
 | |
2ZFE
 
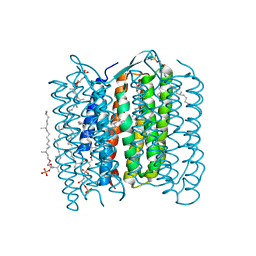 | | Crystal structure of bacteriorhodopsin-xenon complex | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, ... | | Authors: | Kouyama, T. | | Deposit date: | 2007-12-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of xenon binding to a hydrophobic cavity on the proton pumping cycle in bacteriorhodopsin
J.Mol.Biol., 384, 2008
|
|
3WQJ
 
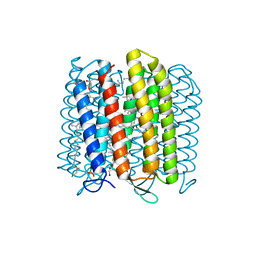 | | Crystal structure of archaerhodopsin-2 at 1.8 angstrom resolution | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, ... | | Authors: | Kouyama, T. | | Deposit date: | 2014-01-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of archaerhodopsin-2 at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3A7K
 
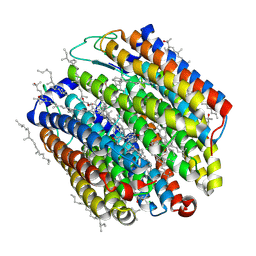 | | Crystal structure of halorhodopsin from Natronomonas pharaonis | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Kouyama, T. | | Deposit date: | 2009-09-27 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Light-Driven Chloride Pump Halorhodopsin from Natronomonas pharaonis.
J.Mol.Biol., 396, 2010
|
|
2BDY
 
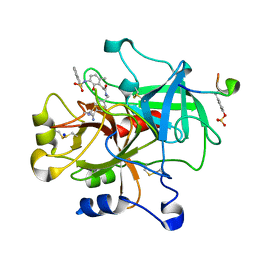 | | thrombin in complex with inhibitor | | Descriptor: | Hirudin IIIB', N-(4-CARBAMIMIDOYL-BENZYL)-2-[2-HYDROXY-6-METHYL-3-(NAPHTHALENE-1-SULFONYLAMINO)-PHENYL]-ACETAMIDE, SODIUM ION, ... | | Authors: | Xue, Y. | | Deposit date: | 2005-10-21 | | Release date: | 2006-10-24 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Phenolic P2/P3 core motif as thrombin inhibitors--design, synthesis, and X-ray co-crystal structure.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2D80
 
 | | Crystal structure of PHB depolymerase from Penicillium funiculosum | | Descriptor: | PHB depolymerase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2D81
 
 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2E0C
 
 | |
2E5M
 
 | |
2N42
 
 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N46
 
 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
