5Y3B
 
 | |
4ZID
 
 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 obtained from Escherichia coli | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Yamanaka, M, Nagao, S, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells
Sci Rep, 6, 2016
|
|
4YSV
 
 | |
2JZ4
 
 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
1GIQ
 
 | | Crystal Structure of the Enzymatic Componet of Iota-Toxin from Clostridium Perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, IOTA TOXIN COMPONENT IA | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1GCP
 
 | | CRYSTAL STRUCTURE OF VAV SH3 DOMAIN | | Descriptor: | VAV PROTO-ONCOGENE | | Authors: | Nishida, M, Nagata, K, Hachimori, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel recognition mode between Vav and Grb2 SH3 domains.
EMBO J., 20, 2001
|
|
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | Authors: | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5B0O
 
 | | Structure of the FliH-FliI complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | Authors: | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5AWI
 
 | | Domain-swapped cytochrome cb562 dimer | | Descriptor: | HEME C, SULFATE ION, Soluble cytochrome b562, ... | | Authors: | Miyamoto, T, Kuribayashi, M, Nagao, S, Shomura, Y, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Domain-swapped cytochrome cb562 dimer and its nanocage encapsulating a Zn-SO4 cluster in the internal cavity
Chem Sci, 2015
|
|
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | Descriptor: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
3ATA
 
 | |
5XLE
 
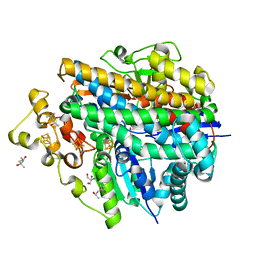 | | Crystal structure of anaerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
3ATE
 
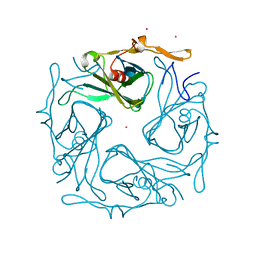 | |
4E9S
 
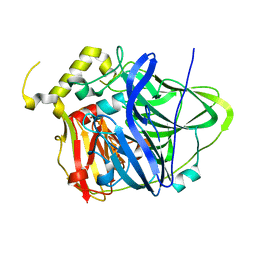 | | Multicopper Oxidase CueO (data5) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site.
J.Mol.Biol., 373, 2007
|
|
5Y0L
 
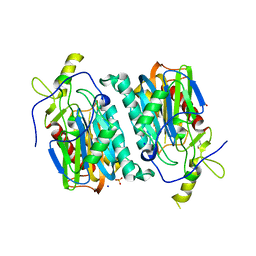 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G/H130Y mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SODIUM ION, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.385 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72. | | Descriptor: | CHLORIDE ION, Endotype 6-aminohexanoat-oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-07 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYS
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122V mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
4E9Q
 
 | | Multicopper Oxidase CueO (data2) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site.
J.Mol.Biol., 373, 2007
|
|
5Y0M
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D36A/D122G/H130Y/E263Q mutant | | Descriptor: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYQ
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122K mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y4N
 
 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
3KLJ
 
 | | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-dependent dehydrogenase, NirB-family (N-terminal domain) | | Authors: | Nishikawa, K, Shomura, Y, Kawasaki, S, Niimura, Y, Higuchi, Y. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of NADH:rubredoxin oxidoreductase from Clostridium acetobutylicum: a key component of the dioxygen scavenging system in obligatory anaerobes.
Proteins, 78, 2010
|
|
7WAF
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAC
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) | | Descriptor: | Cyanophycin synthase | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAD
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS | | Descriptor: | Cyanophycin synthase, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
